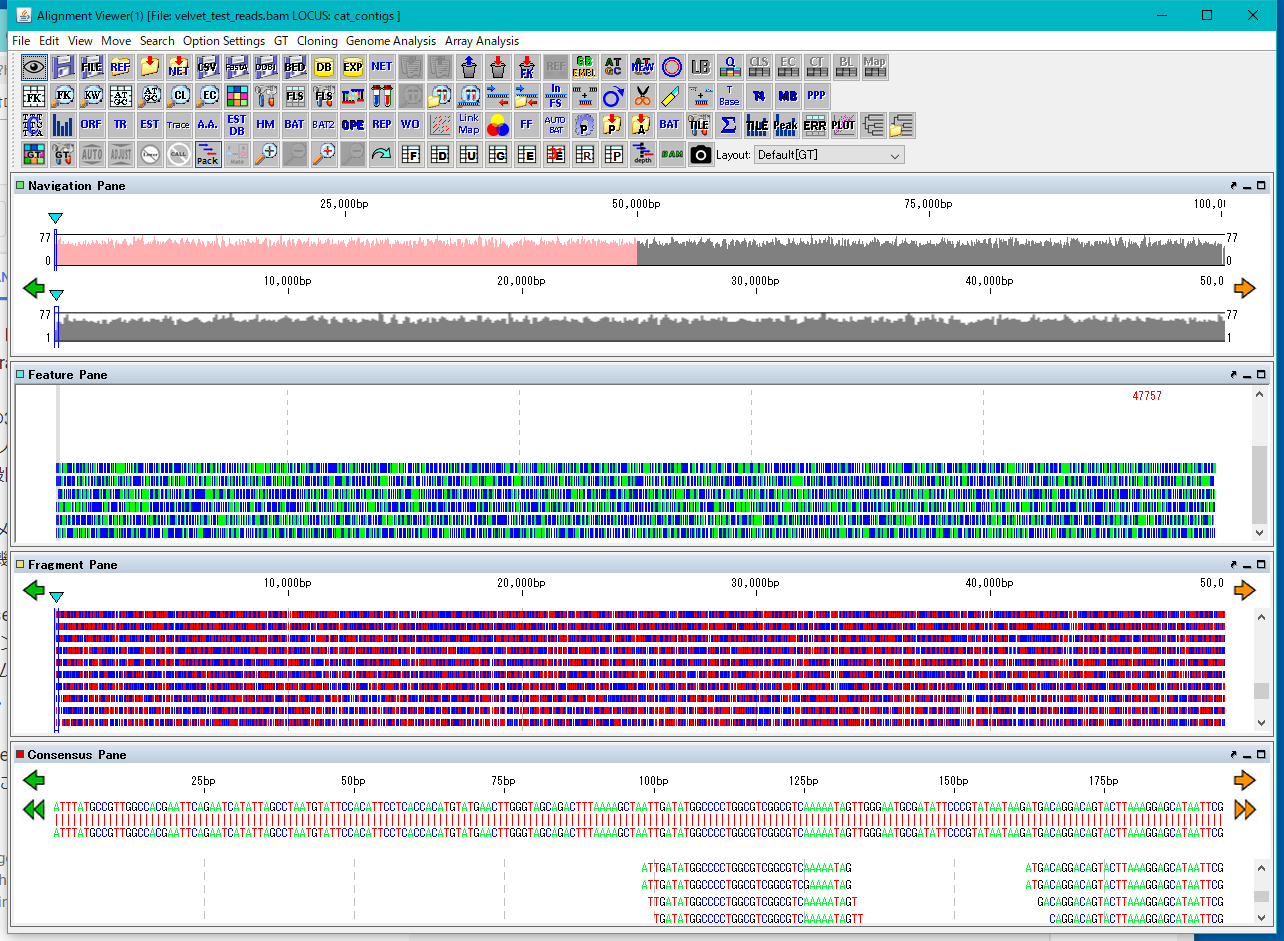
GT's alignment viewer can be launched from TREE Pane in the GT main window.
In order to start up, it is necessary that Analysis nodes that have undergone primary analysis such as importing, assembly, mapping, etc. are registered and analysis data exists in them.
In order to launch the alignment viewer, it is further necessary that these analysis data are saved in BAM format.
As assembly results are output / saved in afg format after the execution is completed, file conversion from afg format to BAM format is required before starting the alignment viewer.
This operation can be done on the node.
Implemented Editioin:
GenomeTraveler![]()
Operation
- The training data is used here for explanation.
- Right-click on the root node in the Tree pane of the GT main window.
- The menu will be displayed.
- Select Expand All.
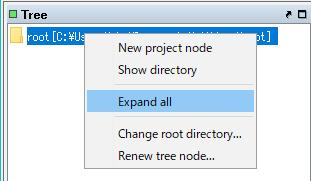
- Tree will be expanded.
- If the Analysis node is not expanded, double-click it or click the "+" in front of the node.
- All nodes will be expanded.
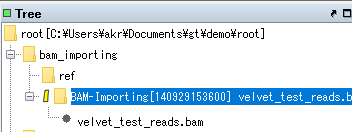
- If the file extension is bam, right click on it.
- The menu will be displayed.
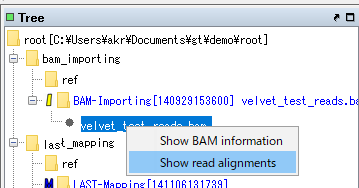
- Select "Show Read Alignment ...".
- If data conversion is required, a conversion execution confirmation message is displayed.
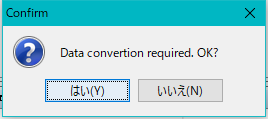
- Click "Yes (Y)".
- The conversion process and launcher process are started.
- While the conversion is in progress, a progress message is displayed.
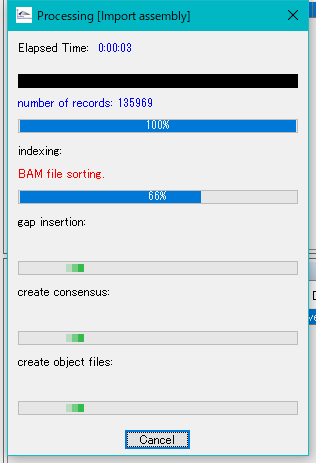
- When the conversion is completed, the alignment viewer is lauched.
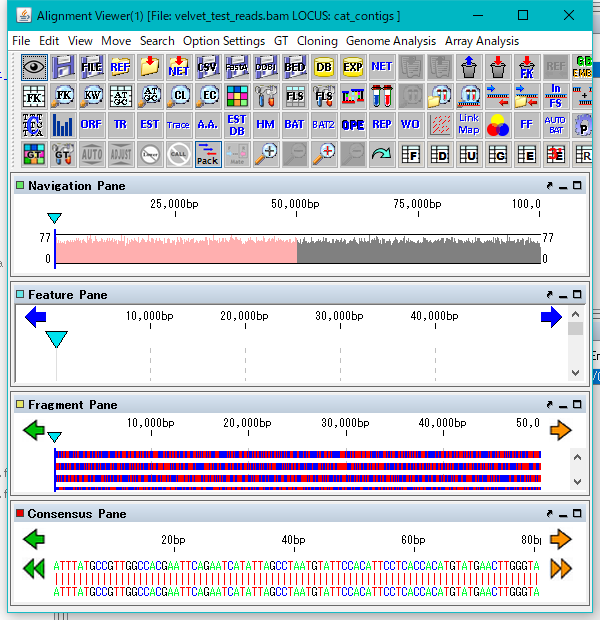
- Resize the launched alignment viewer to the appropriate size.