In this tab pane, you can change the parameter settings for PCR Primer design.
Due to the function to activate primer design, there are additional parameter settings. These are described in the feature page to launch.
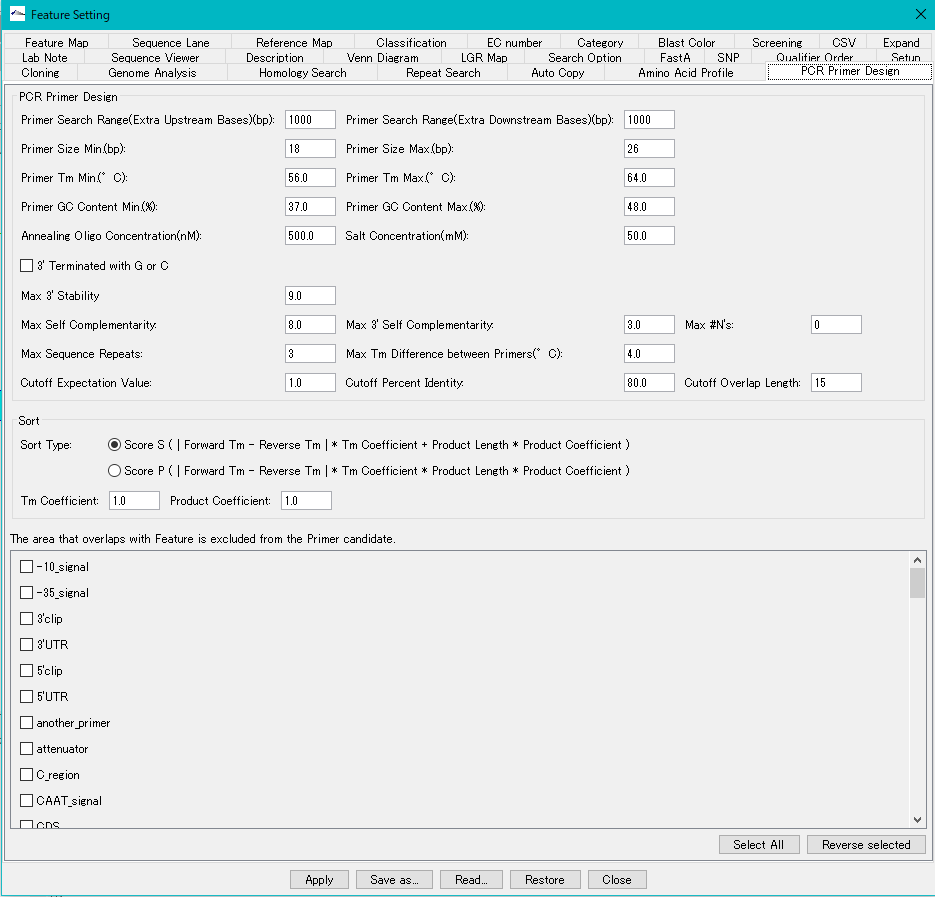
PCR Primer Design section
Primer Search Range (Extra Upstream Bases text field: Specify the Priming Site search range Specify the search range upstream of the selection area as the number of bases Enter a positive integer.
Primer Search Range (Extra Downstream Bases text field: Specify the Priming Site search range Specify the range to be searched downstream of the selection area as the number of bases Enter a positive integer.
Primer Size Min. Text field: Enter the minimum value of primer base length. Enter a positive integer.
Primer Size Max. Text field: Enter the maximum value of primer base length. Enter a positive integer.
Primer Tm Min. Text field: Enter the minimum value of primer Tm. Enter a positive real number.
Primer Tm Max. Text field: Enter the maximum value of the primer Tm. Enter a positive real number.
Primer GC Content Min. Text field: Enter the minimum value of the primer GC content. Enter a positive real number.
Primer GC Content Max. Text field: Enter the maximum value of the primer GC content. Enter a positive real number.
Annealing Oligo Concentration (nM) text field: Enter the annealing oligo concentration. Enter a positive real number. Normally you do not need to change the setting.
Salt Concentration (mM) text field: Enter the salt concentration. Enter a positive real number. Normally you do not need to change the setting.
3 'Terminated with G or C check box: If checked, limit the 3' terminal base of the primer to G or C.
Max 3 'Stability text field: Enter the maximum stability of the 3' end 5 bases of the candidate primer. Enter a positive real number.
Max Self Complementarity text field: Excludes candidate primer sequences that have self-complementary sequences greater than or equal to the specified number of bases.
Max 3 'Self Complementarity text field: Excludes candidate primer sequences that have self-complementary sequences at the 3' end with a specified number of bases or more.
Max # N's text field: Enter the number of N bases allowed in the Priming Site. Enter zero or a positive integer.
Max Sequence Repeats text field: Specify the maximum number of repetitions of the same base. Enter a positive integer.
Max Tm Difference between Primers text field: Specify the maximum Tm difference between both primers. Enter a positive real number.
Cutoff Expectation Value text field: Specify the maximum value of e-value that the site exhibiting homology other than the candidate Priming Site has. If it is more than the maximum value, it is excluded from the candidate.
Cutoff Percent Identity text field: A site showing homology other than the candidate Priming Site
Specify the maximum value of Percent Identity. If it is more than the maximum value, it is excluded from the candidate.
Cutoff Overlap Length text field: Specify the maximum value of Overlap Length of the part showing homology other than the candidate Priming Site. If it is more than the maximum value, it is excluded from the candidate.
Sort section: Set the display order of the designed PCR Primer set list
Sort Type
Score S toggle radio button: Use this expression.
Score P toggle radio button: Use this formula.
Tm Coefficient text field: Enter the coefficient of Tm to be assigned to the calculation formula of the score.
Product Coefficient text field: Enter the coefficient of Product to be assigned to the calculation formula of the score.
The Area that Overlaps with Feature is excluded from the Primer Candidates Section
A list of all feature keys is displayed. There is a check box for selection. Primers with a Priming Site at a location with the checked feature key are excluded.
Select All button: Click to check all lines in the list.
Reverse Selected button: When clicked, the currently selected line is unchecked, and the unselected line is selected.
For Feature Setting common operation buttons, please click here.