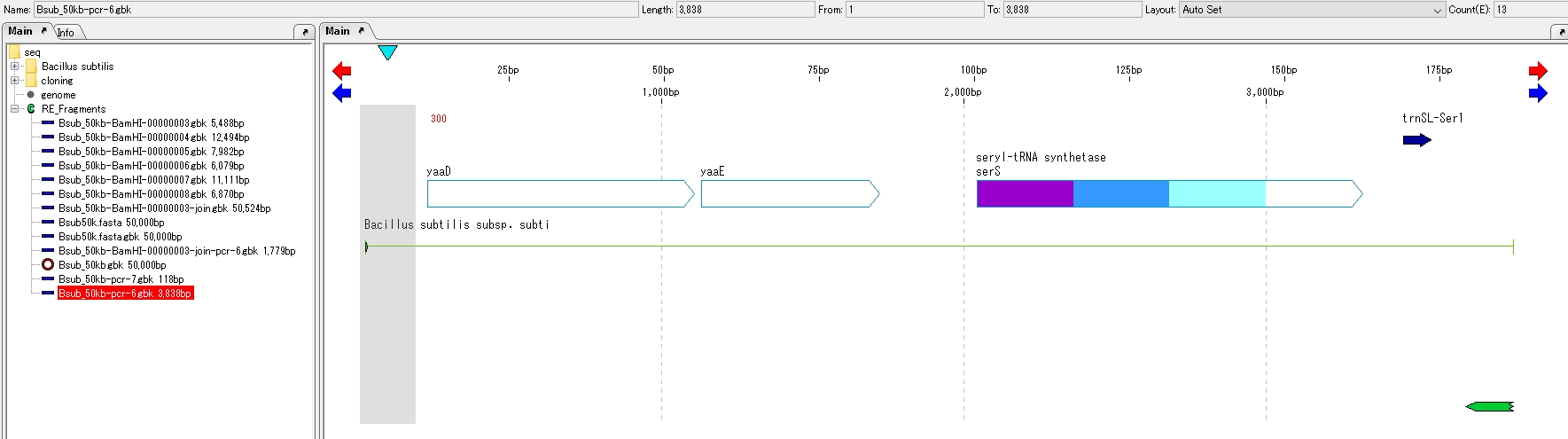
Operation
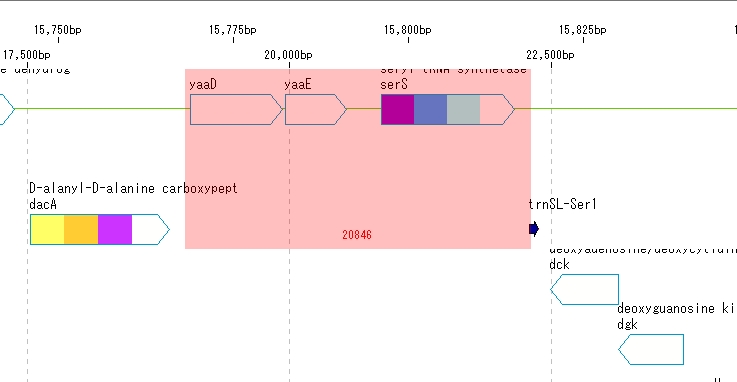
Area designation
- Drag any area on the feature map with the mouse.
- The dragged area is displayed in red.
Primer design
- Click the right mouse button on the feature map.
- A pop-up menu will be displayed.
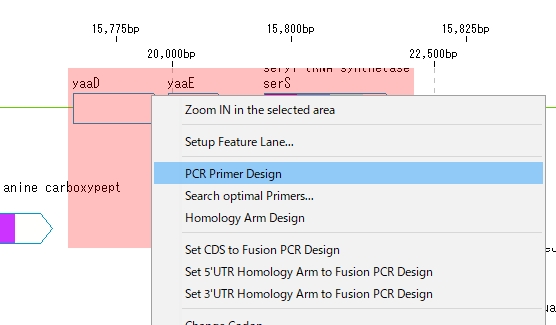
- Select "PCR Primer Design" from the menu.
- The “PCR Primer Design” dialog is displayed.
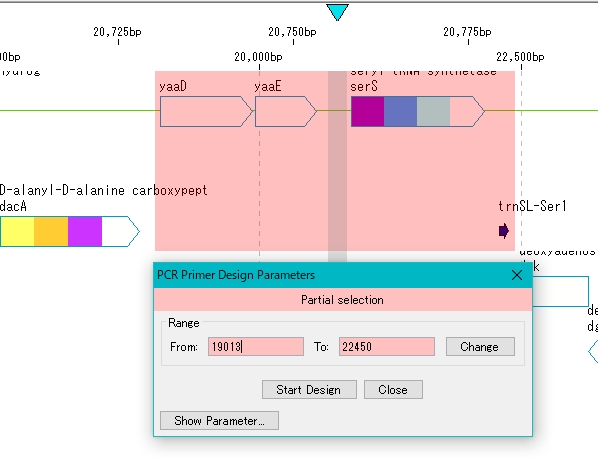
When setting different from the default
- Click Show Parameters in the PCR Primer Design dialog.
- The detailed parameters are displayed.
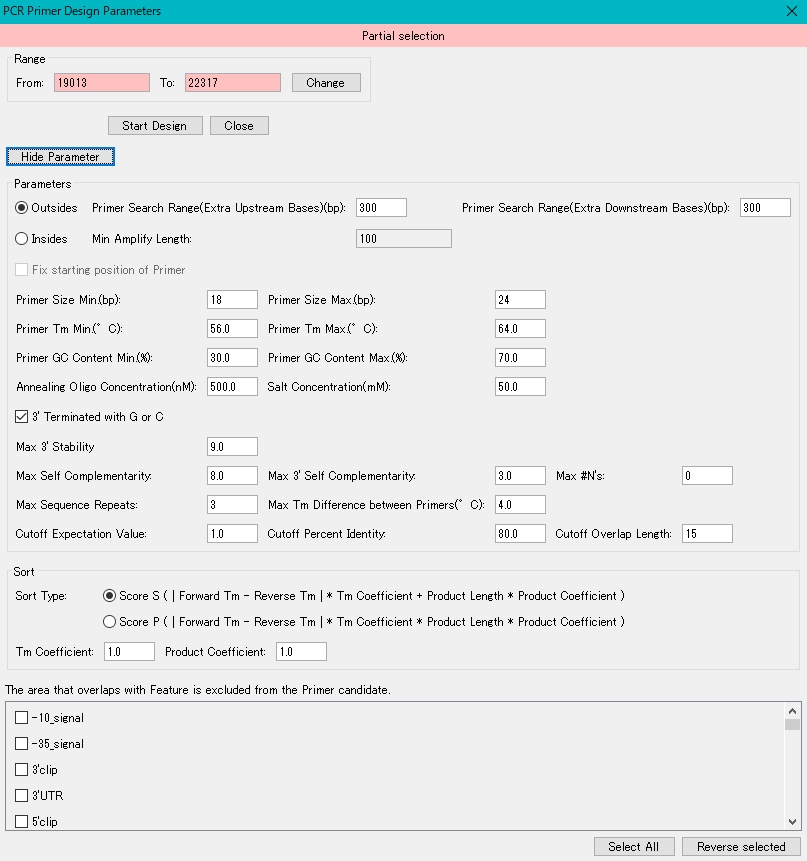
- To change the region to be amplified, enter a value directly in the "From:" and "To:" fields, and click the "Change" button.
- The amplification target area is changed.
- To change how many bases upstream of the amplification region are searched for the primer priming site, enter a number directly in the “Primer Search Range (Extra Upstream Bases) (bp)” field.
- In this case, the primer is searched from the base position of From: up to the number of bases entered in this field.
- Set the same for the downstream side.
- For the range of the base length of the primer, enter the minimum base length and the maximum base length directly in the “Primer Size Range Max.” Field.
- For the Tm range of the primer, enter the minimum Tm temperature (° C) and maximum Tm temperature (° C) directly in the “Tm Range Min.” And “Tm Range Max.” Fields.
- For the range of the GC content of the primer, enter the minimum GC content (%) and the maximum GC content (%) directly in the "GC Range Min." And "GC Range Max." Fields.
- To specify the annealing oligo concentration, enter the concentration (nM) directly in the "Annealing Oligo Concentration" field.
- To specify the salt concentration, enter the circular concentration (nM) directly in the "Salt Concentration" field.
- Indicates the score threshold for judging whether there is primer self-complementarity.
- If the score is less than or equal to the Max SelfarComplementarity field score, the primer is acceptable.
- When detecting repetitive sequences in the primers, specify the maximum base length that will not be recognized as a repetitive sequence in the "Max Sequence Repeats" field.
- That is, if there are two or more identical sequences with a base length longer than this base length, they will not be used as primers.
- Specify the maximum Tm difference between primer pairs by directly entering it in the "Max Tm Differenece" field.
- Three parameters are prepared to check whether there is a priming site in the target DNA region.
- For the check, Blast1 is used, so the parameters are the same as those of Blast.
- “Cutoff Expectation Value” indicates the expected value that is the detection threshold.
- "Cutoff Percent Identity" indicates the threshold value of what percentage or more of the primer base lengths match.
- “Cutoff Overlap Length” indicates the minimum number of base overlaps between the primer and the priming DNA sequence.
- Click the "Set" button.
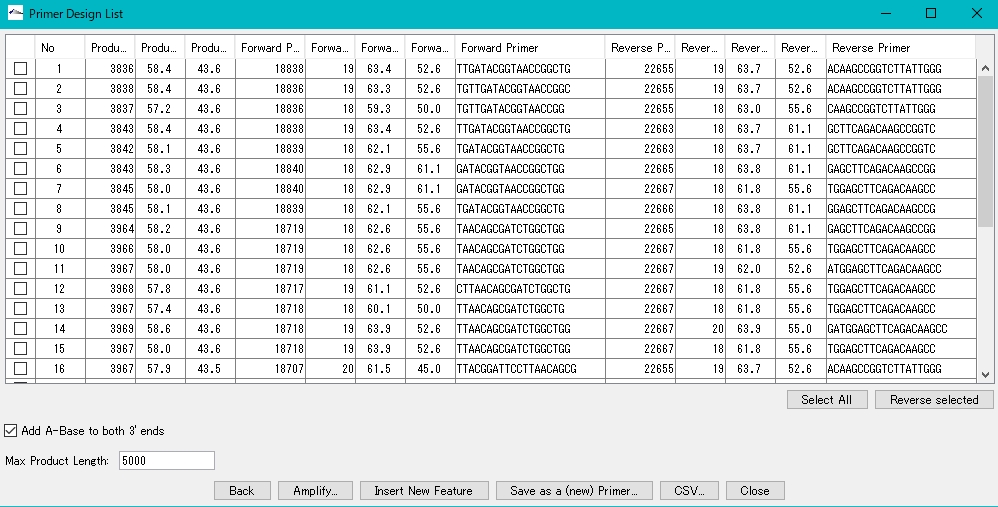
- A window listing the optimal PCR Primer Set appears.
- An error message is displayed if the optimal Primer Set cannot be designed.
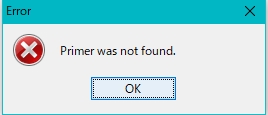
- If you cannot design the Primer, change the design parameters and redesign.
Confirmation of amplification region
- Clicking on each line of the Primer Set List displays the amplification region corresponding to that Primer Set in color.
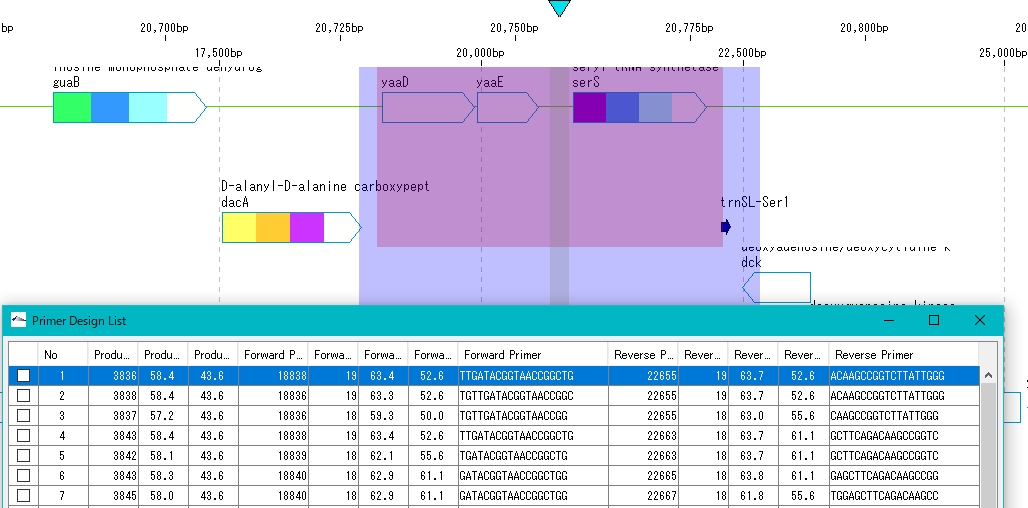
Perform PCR amplification
- Check the box of Primer Set to be used from Primer Design {List}.
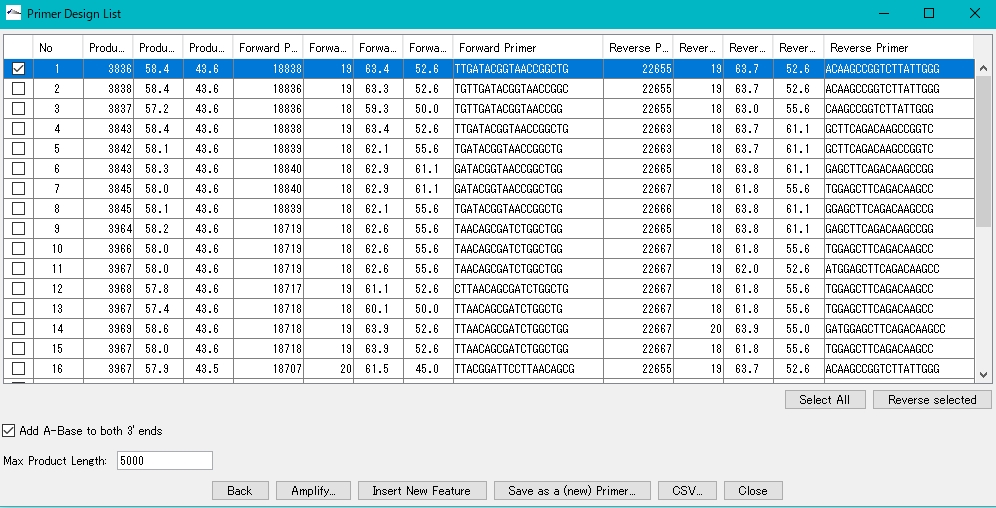
- Click Amplify.
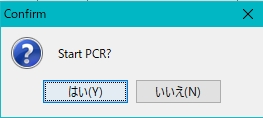
- The confirmation message “Start PCR” appears.
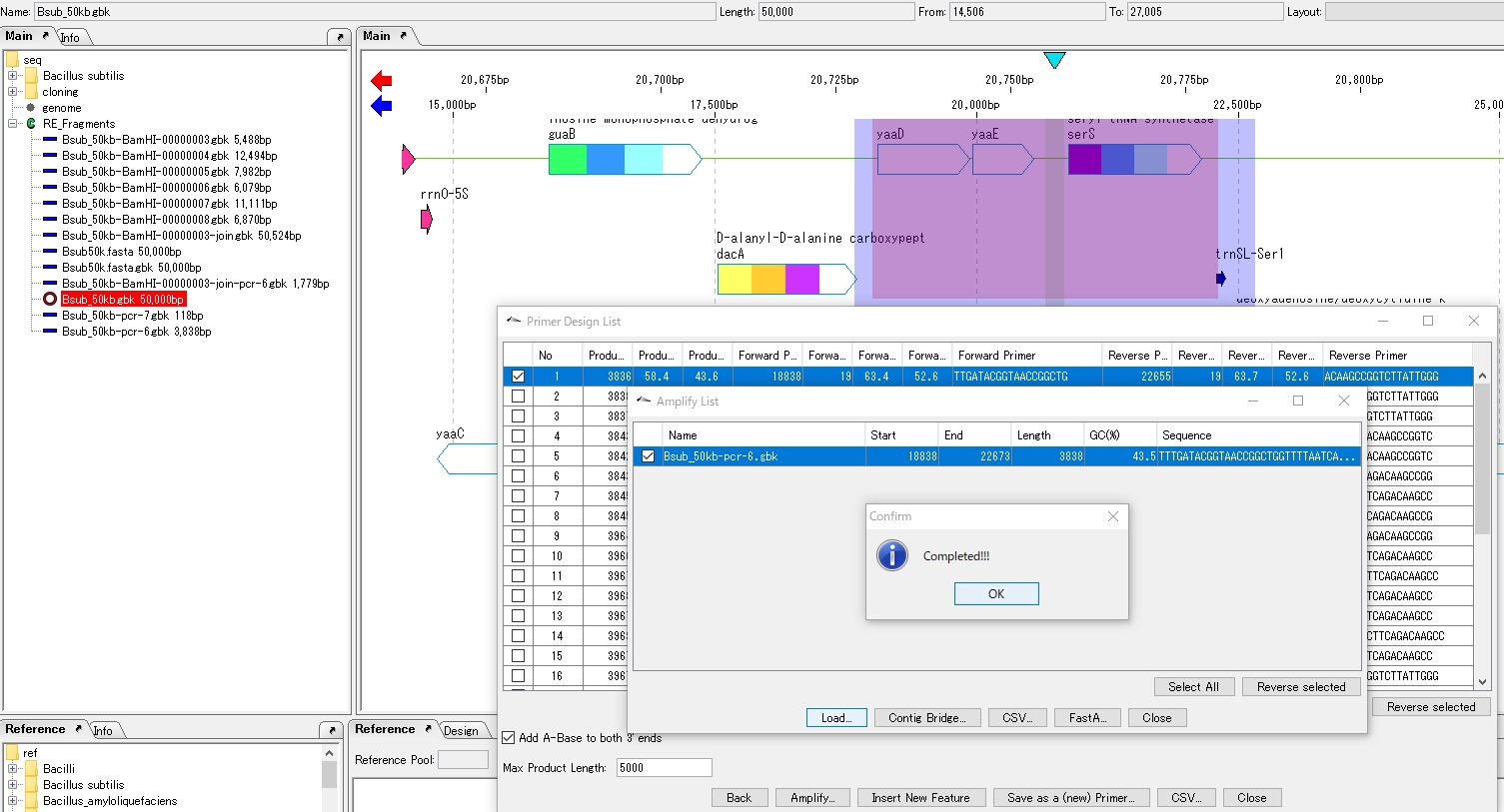
- The Amplify List window appears.The list lists the amplification products.
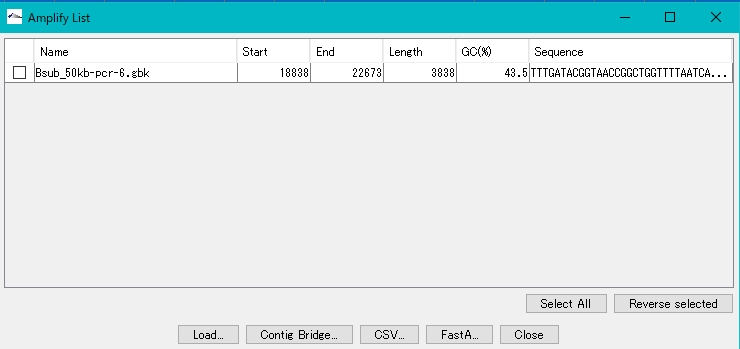
Loading amplification products
- To add an amplification product to the main directory, check the checkbox for that product.
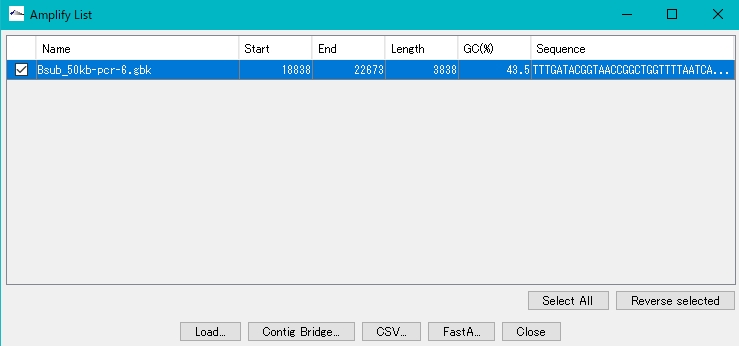
- Click Load.
- The Load is executed, and when it is completed, a completion message is displayed.
- The generated sequence of the PCR product is added to the current main directory.
Browse amplification product sequences
- Click the sequence of the PCR product added to the current main directory.
- The feature map of the PCR product is displayed.