Linearize circular vector with restriction enzyme
Operation
- Load the DNA sequence of a circular vector into the current main directory.
- Load the sample sequence "pColdV.gbk" as an example.
- The sequence loaded in the main feature map is displayed.
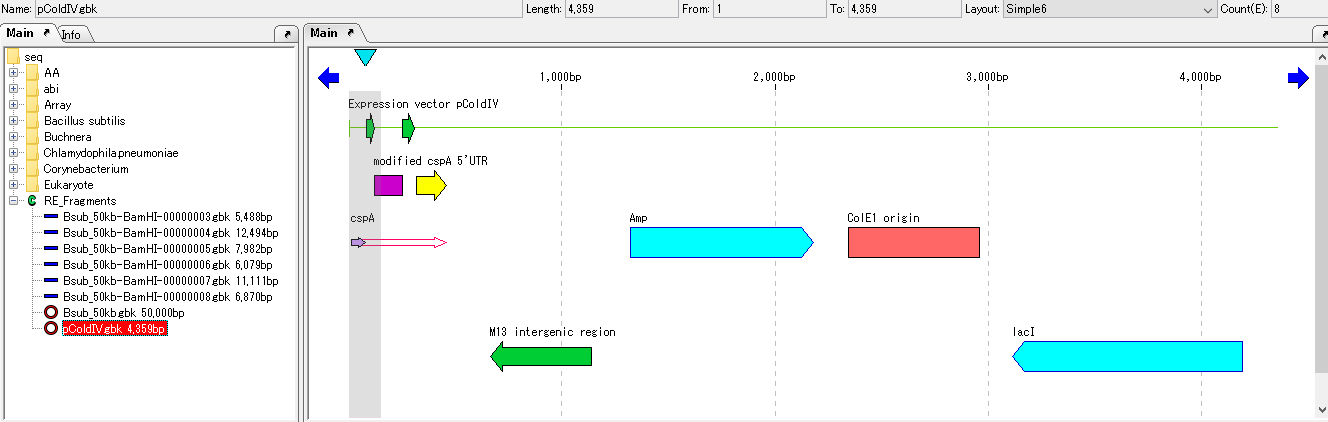
- Find a restriction enzyme that cleaves this vector in only one place.
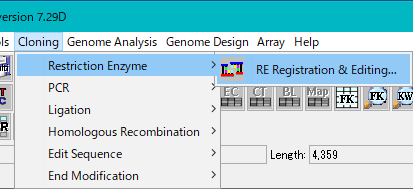
- Click the "Cloning -> Restriction Enzyme -> RE Registration & Editing ..." button from the menu.
- The "Enzyme Selection" dialog is displayed.
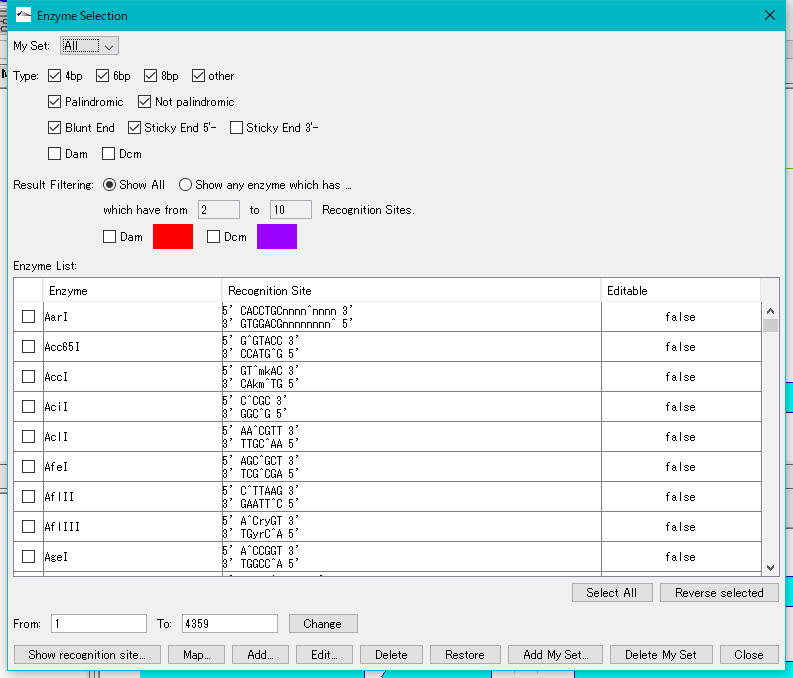
- In the "Select Enzyme" section, in the "Result" field, check "Any Enzyme".
- Enter 1 in the "which have from" and "To" fields.
- Click "Select All". All restriction enzymes are checked.
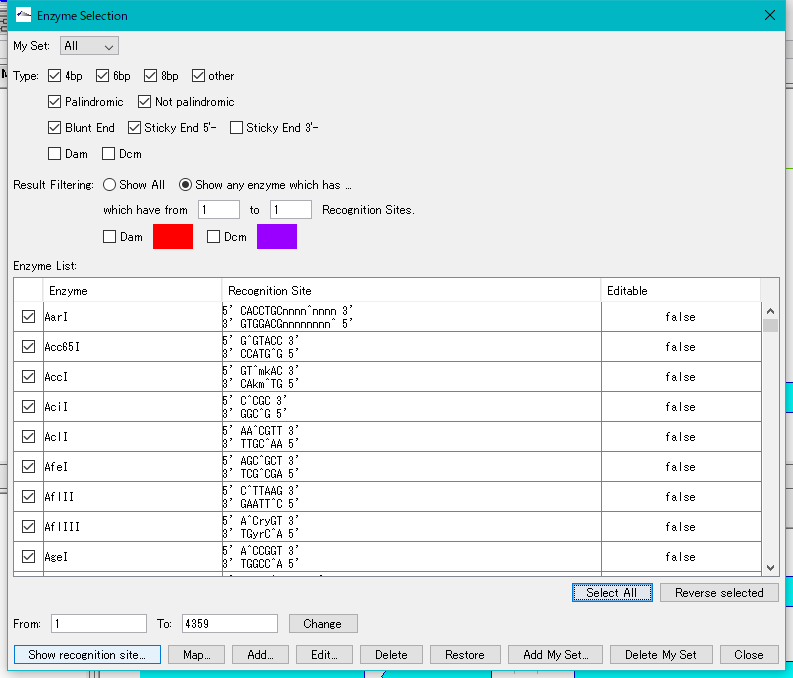
- Click "Show Recognition Site ...".
- The "Recognition Site" dialog is displayed.
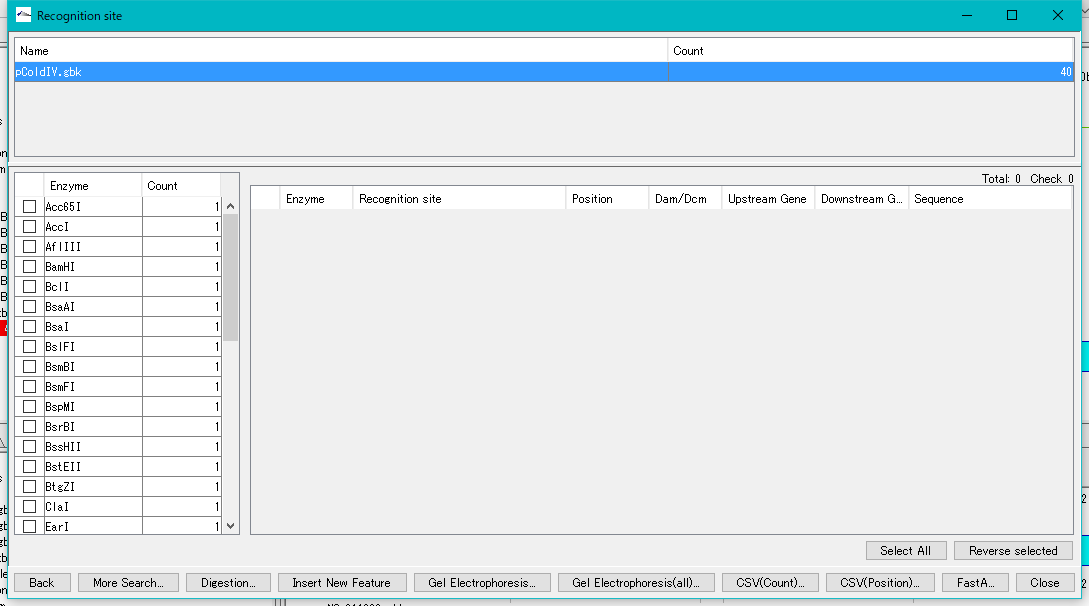
- For example, you know that BamHI disconnects MCS in only one place, so check BamHI.
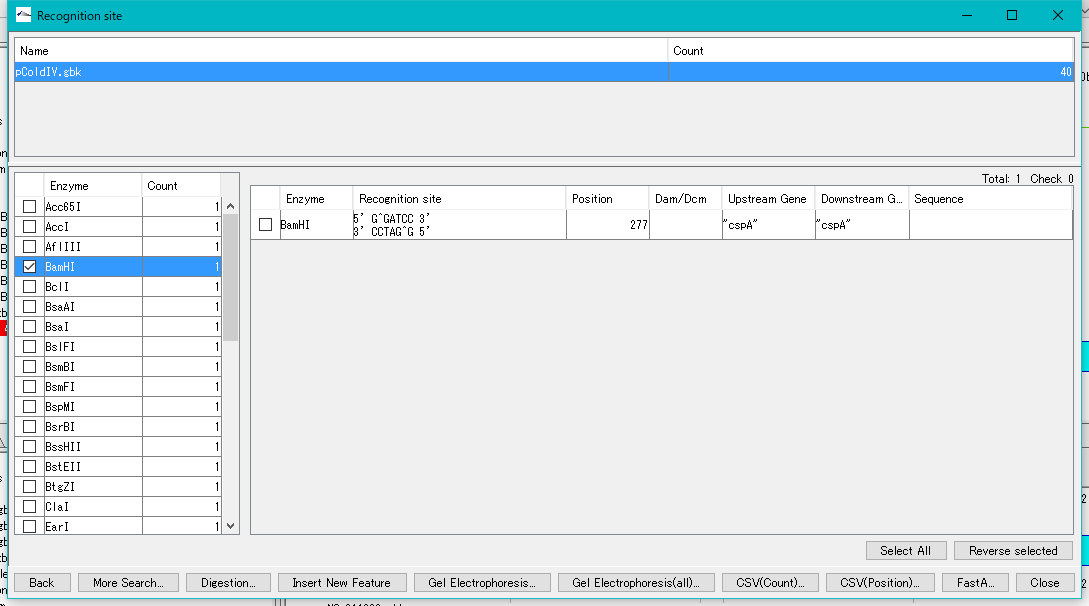
- The recognition site is displayed in the right pane.
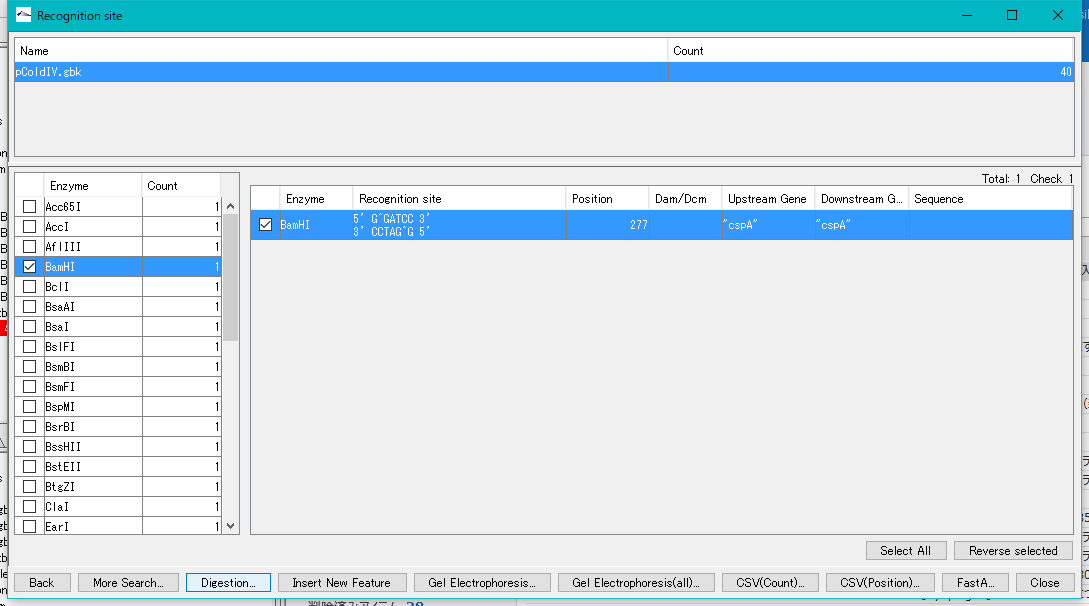
- Check the BamHI in the right pane and click the "Digestion" button.
- A confirmation message will be displayed.
- Click "Yes (Y)".
- The "Digestion list" window will be displayed.
- Only one digested fragment is listed.
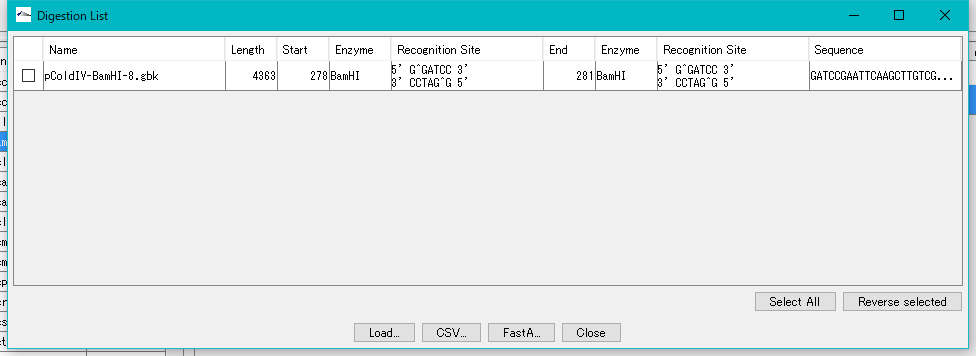
- Check the fragment and click the "Load" button.
- A "Completed !!" completion message is displayed.
- Click "OK".
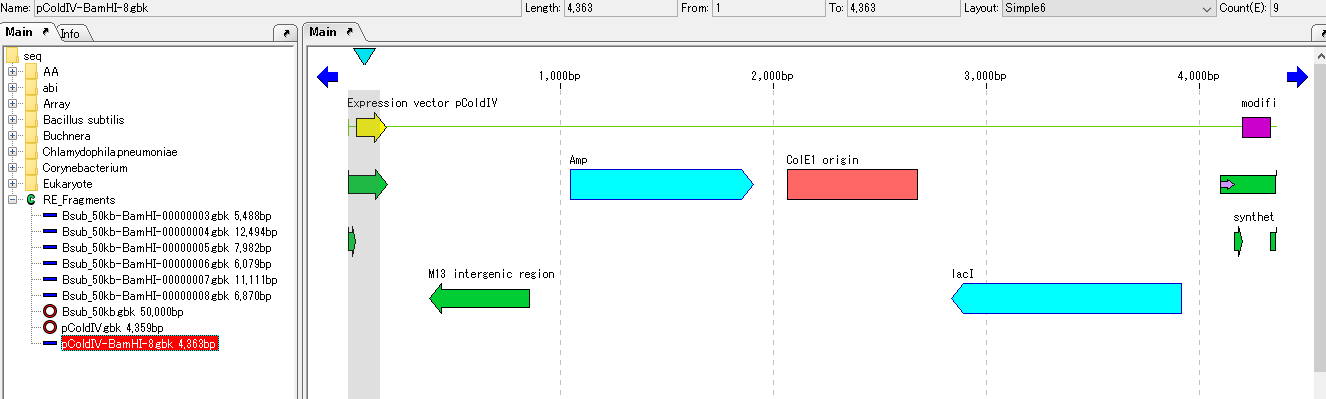
- The vector sequence opened with the main feature map is loaded.
- Linearized DNA has end points for horizontal scrolling.
- Click "Cloning -> Ligation -> Simple Ligation ..." from the menu.
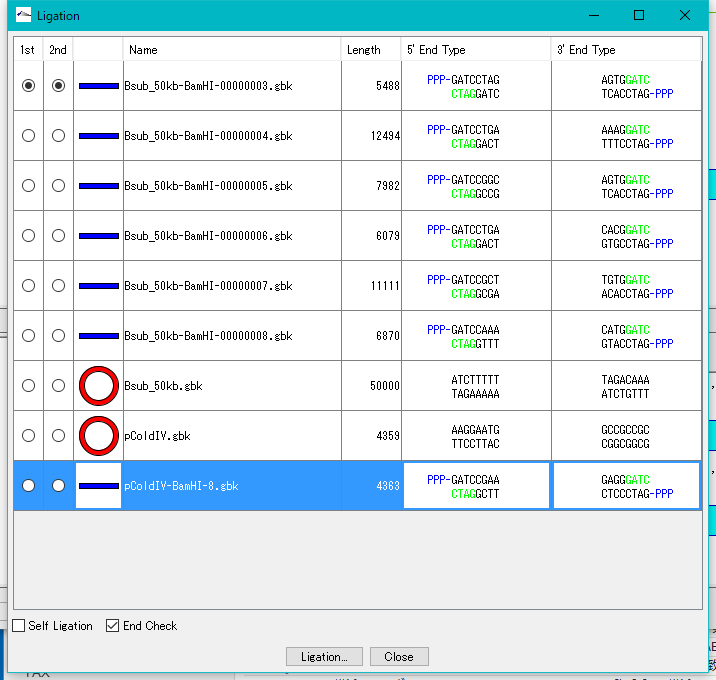
- The Ligation dialog will be displayed.
- There is a vector opened at the bottom of the list, and the shape of both ends is displayed.
- The end shape can also be seen in the sequence lane of the main feature map.
- Scroll horizontally until scrolling stops.