Extract ORF candidates from the prokaryotic genome sequence and register them as new features.
The ORF candidate is judged by the length of the stop codon absence region.
You can set stop codon absence regions (ORF_long, ORF_Middle, ORF_Short) of three kinds of base length ranges.
ORF candidates can be changed to CDS by Translate function.
Operation
- Load the prokaryotic genomic sequence into the main current directory.
- As an example, we will use the sample data Bsub 50 kb.fasta.
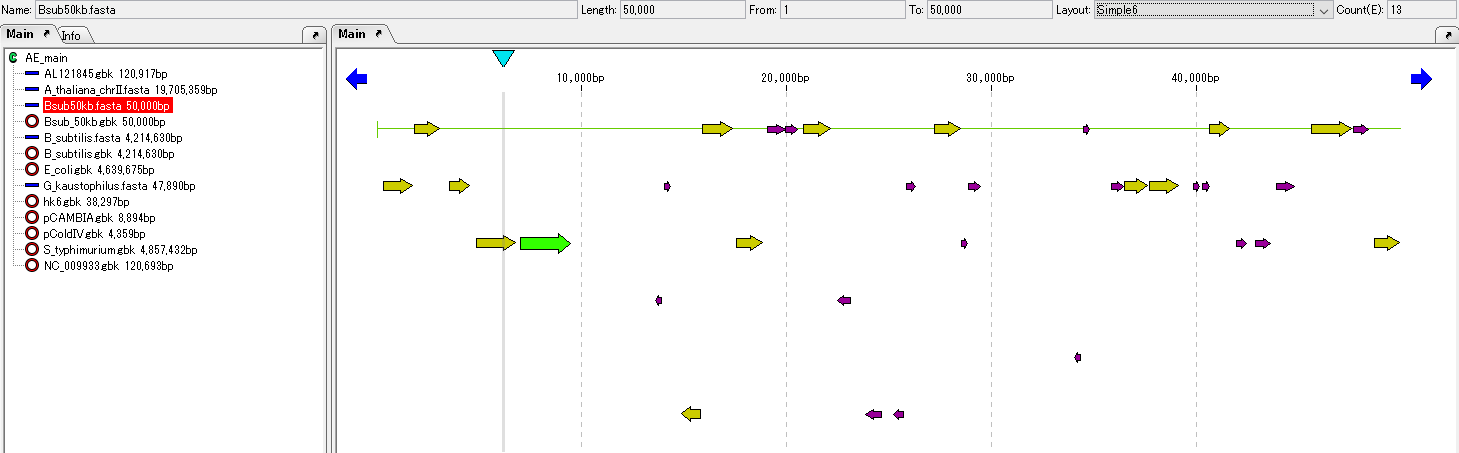
- When loading, it usually becomes the current arrangement and it is displayed in the main feature map.
- If it does not appear in the main feature map, click this array on the main directory.
- Click "Genome Analysis -> ORF -> ORF Extraction" from the menu.
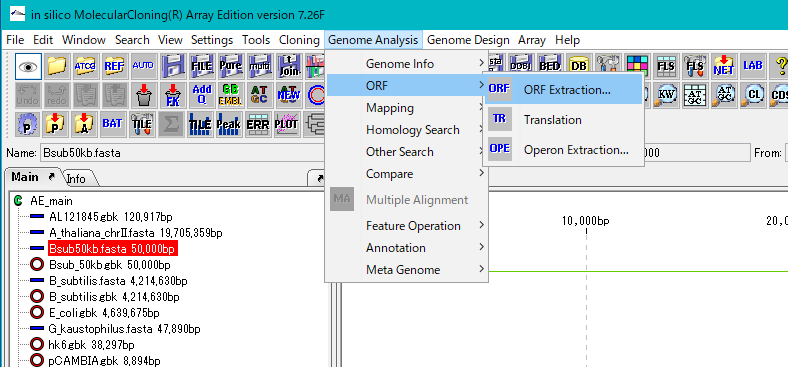
- The "Range Setting" dialog will be displayed.
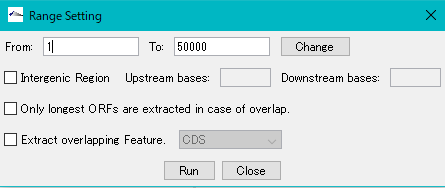
- Check "Only Longest ORFs are Extracted in case of Overlap" checkbox.
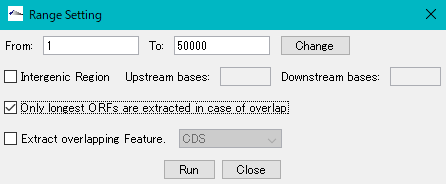
- Click "Run".
- The "Stard ORF Search?" Confirmation message is displayed.
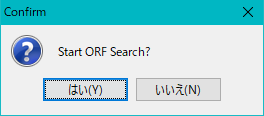
- Click "Yes (Y)".
- When the ORF extraction is completed, the "ORF Site" dialog will be displayed.
- The number of ORF candidate detections by ORF type is displayed in the list on the left.
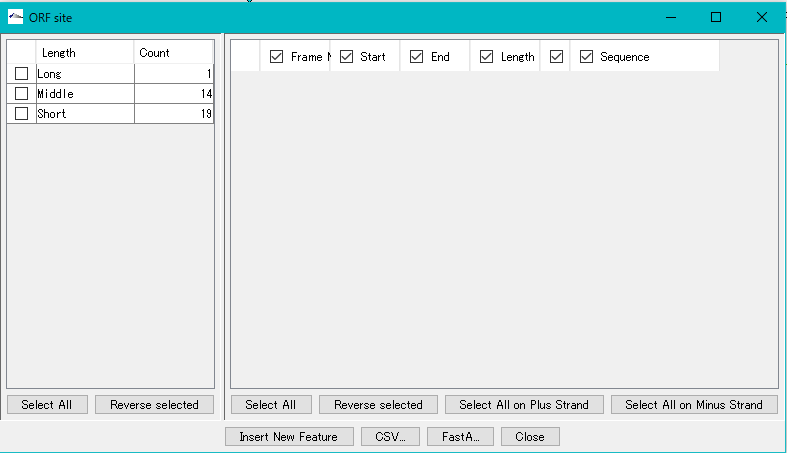
- Check each check box in the list on the left.
- Alternatively, click "Select All" in the lower left.
- The frame No., start base position, end base position, base length, strand and base of each ORF candidate are displayed in the list on the right.
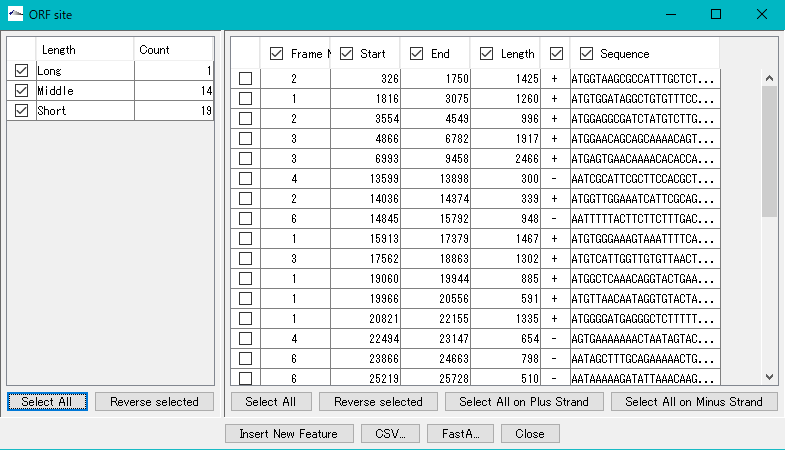
- To register all extracted ORF candidates as features, click "Select All" in the lower right.
- All ORF candidates are checked.
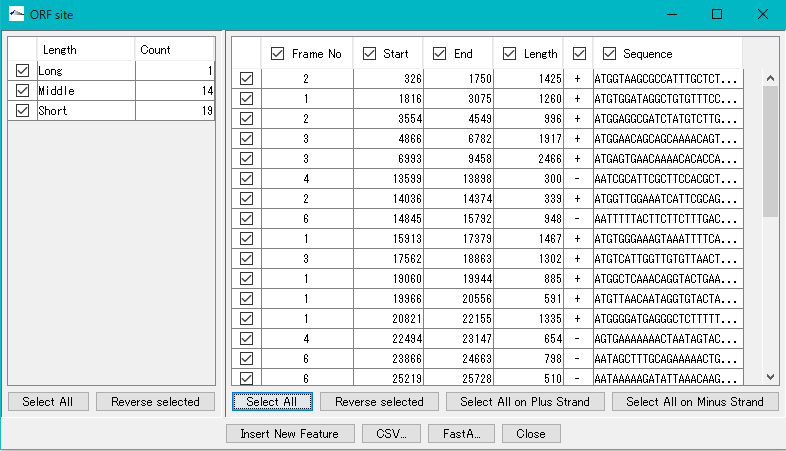
- Click "Insert New Feature".
- The "Insert New Feature?" Confirmation message will be displayed.
- Click "Yes (Y)".
- A "Completed !!!" message will be displayed.
- Click "OK".
- If you do not close the "ORF Site" dialog, you can use it to check the position of each ORF candidate.
- Click one of the ORF candidates in the right list.
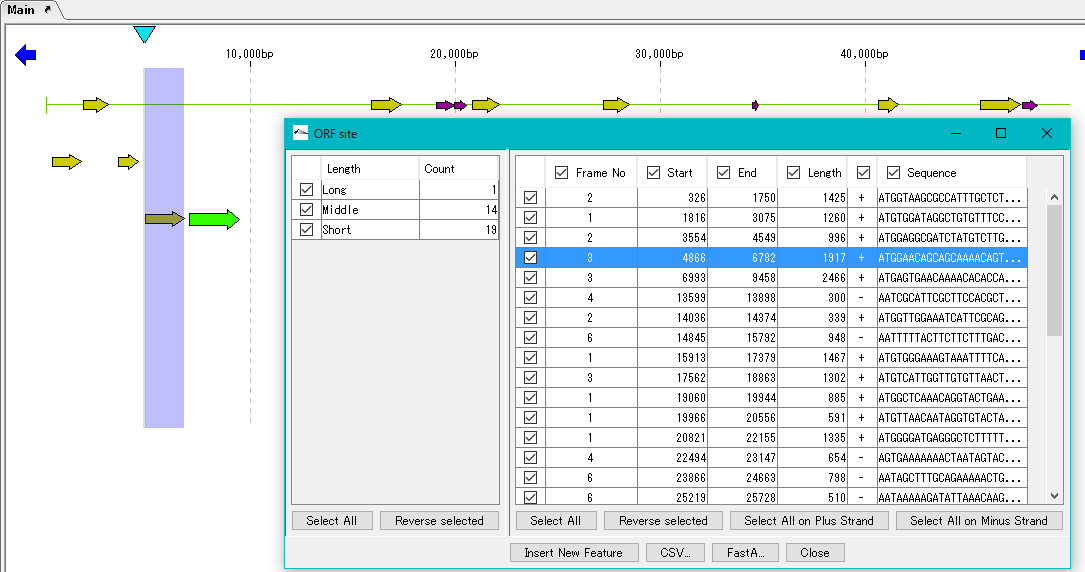
- The position of the corresponding feature in the main feature map is highlighted.
- Click "Close" in "ORF Site" dialog.
- The "ORF Site" dialog closes.
- Feature of registered ORF candidate is displayed on the feature lane of the main feature map.

After this, proceed to "Change the candidate's feature key to CDS and translate amino acids".