In order to register as a CDS the features extracted and registered with "Extract ORF", it is necessary to perform amino acid translation on each and to change the feature key to CDS.
By executing Translation, you can translate amino acids for all ORF candidate features on the current genome sequence, and at the same time change the feature key to CDS.
Operation
Load the genome sequence of ORF candidate extraction and feature registration completed into the main current directory.
When continuing from ORF extraction, it is the current genome sequence.
From the menu click Genome Analysis -> Translation.
The Translation Setting dialog is displayed.
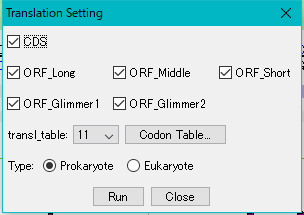
Check all the checkboxes and make sure "transl_table" id is 11 and the type is Prokaryote.
Click "Run".
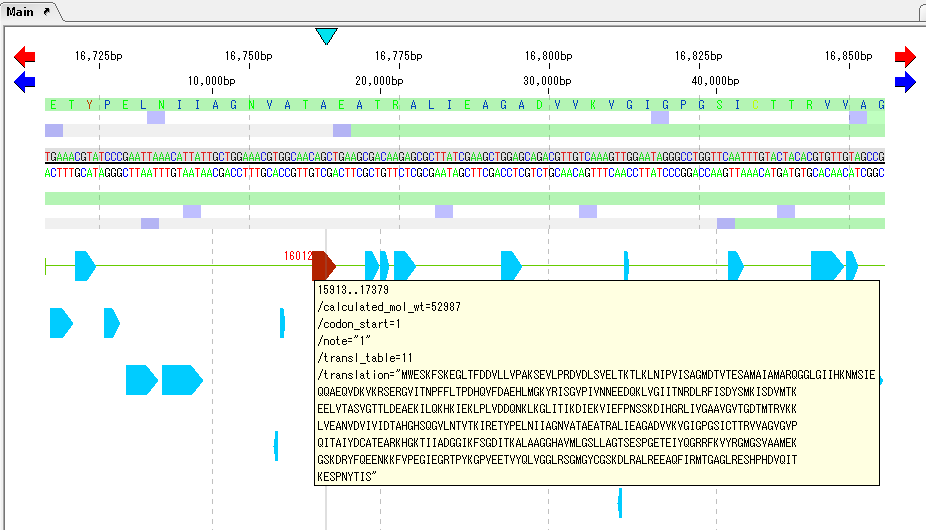
All ORF candidates on the current feature map are converted to CDS and the shape of the feature changes.
The nucleotide sequence corresponding to each CDS feature is amino acid translated according to the specified genetic code table.
Move the mouse over one CDS feature.
A tooltip is displayed and the amino acid sequence is displayed in /translation=.