in silico Assembler is de novo Assembler.
Assemble DNA fragments of 50 bp or more.
Reads from NGS can also be assembled (up to about 1 million reads. For larger size use Velvet etc).
In the preprocessing, limited number of processing, designation of minimum QV, maximum N bases can be limited.
Operation
- Select Tools -> in silico Assembler from the menu.
- If using it for the first time, sample data is not installed. You will be asked if you want to install the sample data.
- The in silico Assembler button tool will be displayed.
- Click Assemble.
- The Assembly execution dialog is displayed.
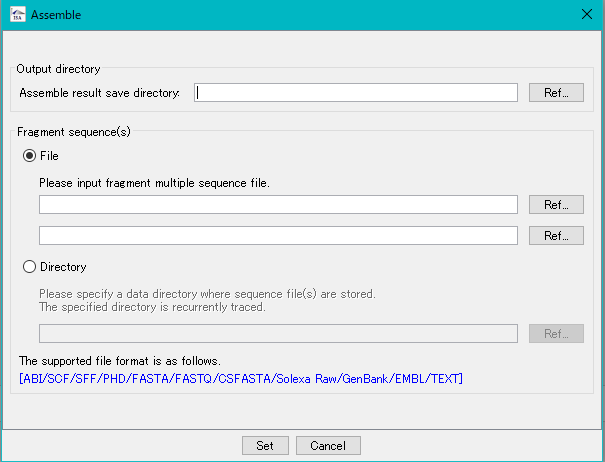
- Specify the save directory of the result.
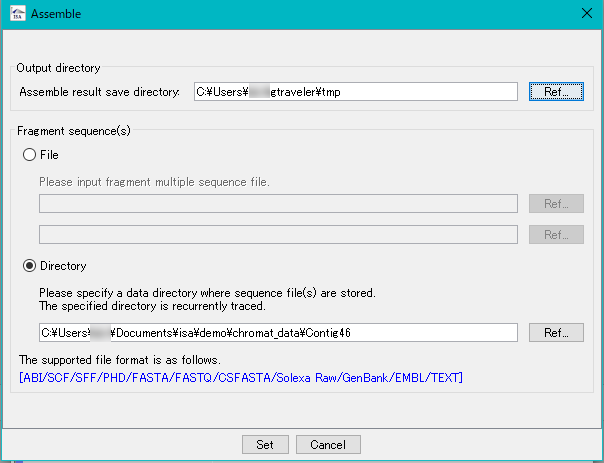
- Specify the DNA fragment files to be assembled.
- The fragment files to be input is specified in one of two ways.
- During assembly execution, a progress message is displayed, so you can grasp the progress of the assembly.
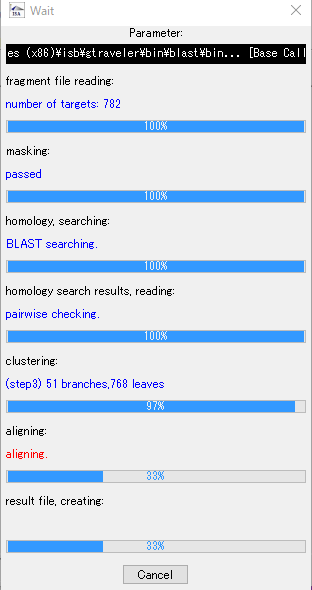
- When complete, a completion message will be displayed.
- Click OK.
- The completion message closes.
Points to be noted
The assembly result is stored in the result storage directory specified before execution.
There is a method of mechanically combining a large number of contig sequences generated as assembly results into one sequence before loading.
Assembly results are usually saved as multiple Contig files, so loading as they are divided into a number of fragment files and loaded.
If there are many fragments, there is a method of specifying all of them and not directly loading, generating and loading a multiple GenBank format file mechanically combining them.
To load assembly results into the main feature map, use IMC's normal sequence loading method.