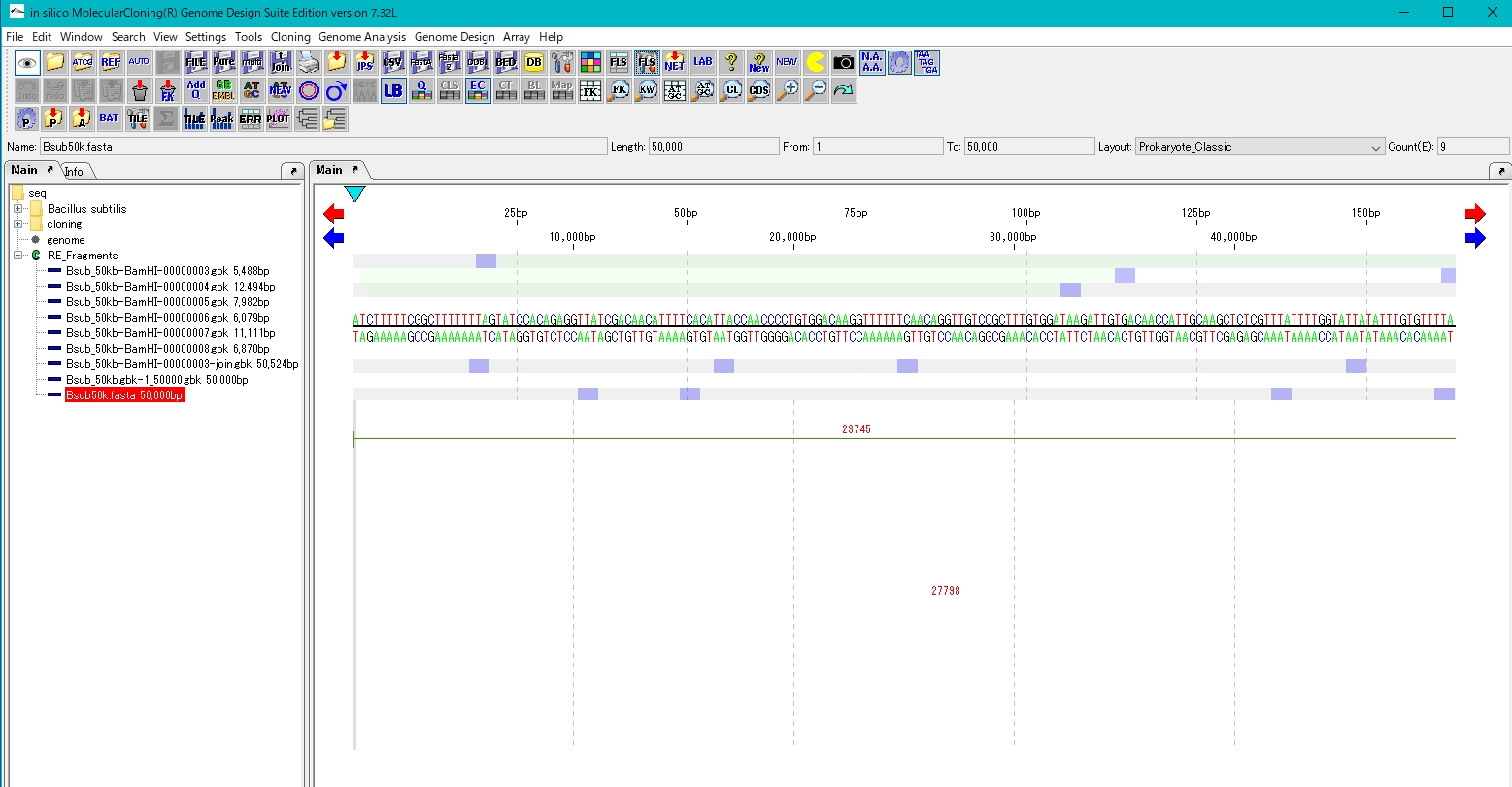
- Map the amino acid sequence on the reference genome sequence and register it as a new feature
- Map the specified amino acid sequence (s) to the reference genome currently displayed as a feature map, and register the hit entry as a new feature (such as mRNA).
- The mapping uses the tBlastN algorithm.
How to operate amino acid sequence mapping
- Read the base sequence of the reference genome and display it on the feature map.
- Click Genome Analysis-> Mapping-> A.A. Sequence Mapping from the menu.
- A.A.Sequence Mapping execution dialog is displayed.
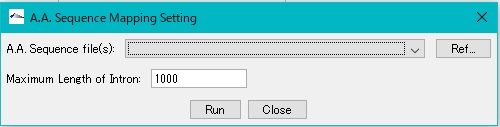
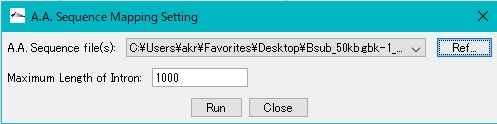
- Click Ref… of A.A.Sequence File (s) and specify the amino acid sequence to be mapped.
- The amino acid sequence that can be specified here is FastA format, GenPept format, etc. Files compressed with gz can also be specified.
- A dialog box for selecting an amino acid sequence appears.
- Specify the amino acid sequence file.
- Click the Run button.
- A confirmation message will be displayed.
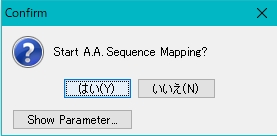
- To change a parameter, click the Parameter button.
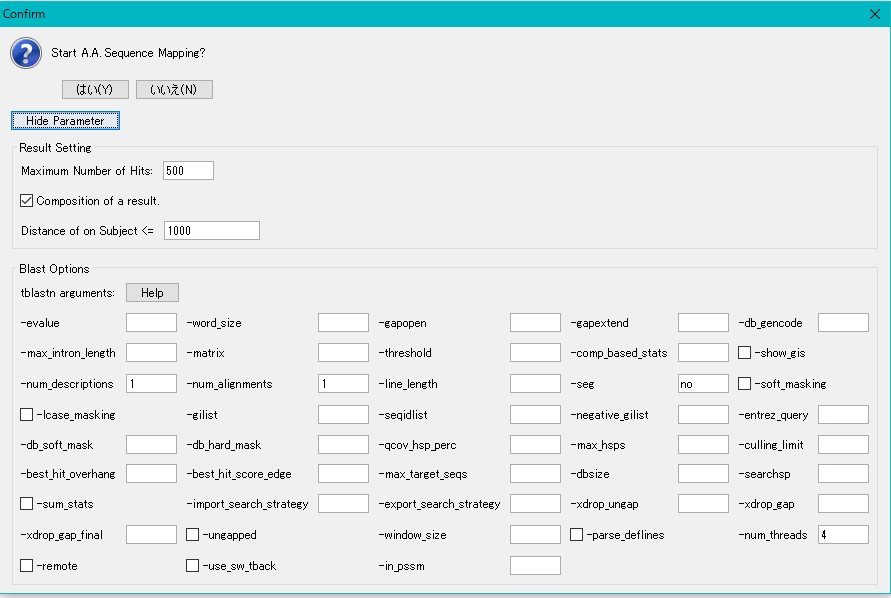
- This parameter is based on NCBI Blast parameter settings.
- Usually use the default parameter settings.
- Click the "Yes" button to start mapping.
- The execution time is proportional to the number and length of amino acid sequences and the base length of the reference genome base sequence.
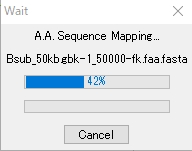
- During execution, a message that indicates the processing progress is displayed.
- When the mapping process is completed, the result list dialog is displayed.
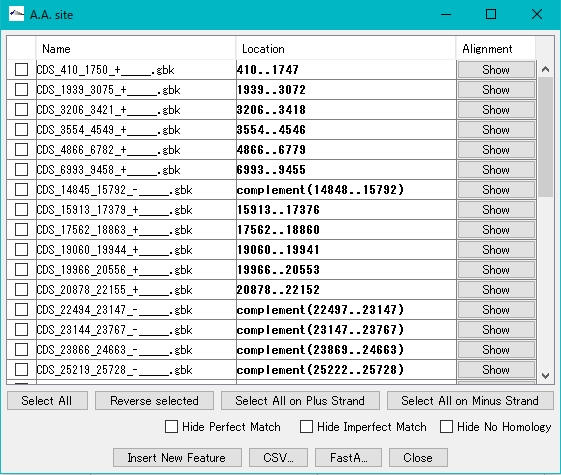
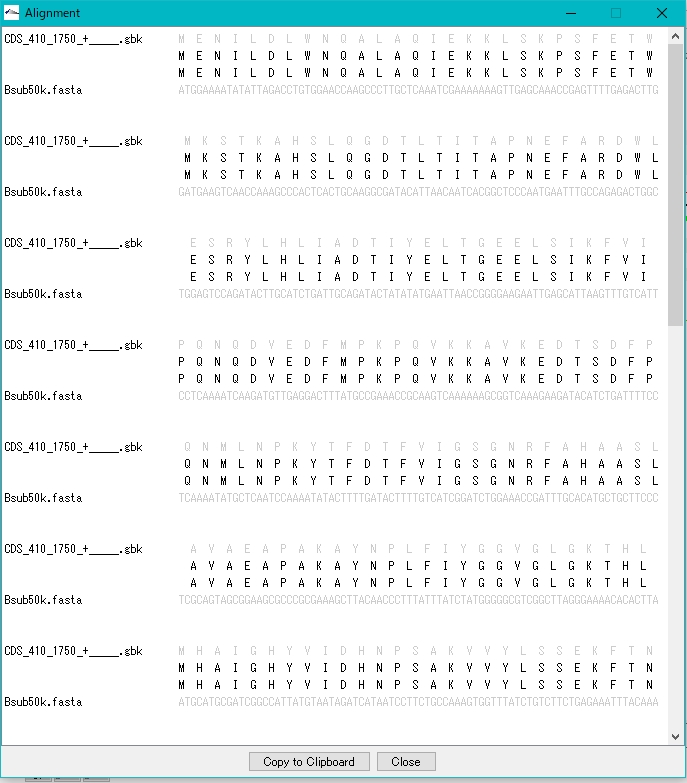
- Clicking on Alignment in each row will bring up a pairwise alignment dialog.
- Register a new feature using the result list dialog.
- Mapping results are classified into three types.
- When the input amino acid perfectly matches the reference genome sequence
- When the input amino acid incompletely matches the reference genome sequence
- When the input amino acid sequence does not match at all
- The former two types of results can be registered as new features on the reference genome. For example, a perfect match sequence can be registered as an mRNA feature, and an
- incomplete match sequence can be registered as a miscRNA feature. It is also possible to specify another feature key such as CDS.
- Click Insert Feature.
- A confirmation message will be displayed.
- Click “Yes”.
- The Feature Setting dialog is displayed.
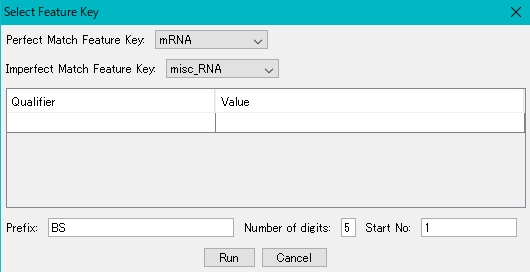
- The same value can be set to any qualifier of the newly registered feature. Also, you can register the prefix specified for Qualifier / locus_id and a serial number that has an arbitrary number of digits and starts with an arbitrary numerical value.

- In the case of eukaryotes with introns, exon and intron regions are identified and registered as features.
- You can limit the maximum base length of an intron.
- Click Run.
- When Feature registration is completed, a completion message is displayed.
- Click OK.
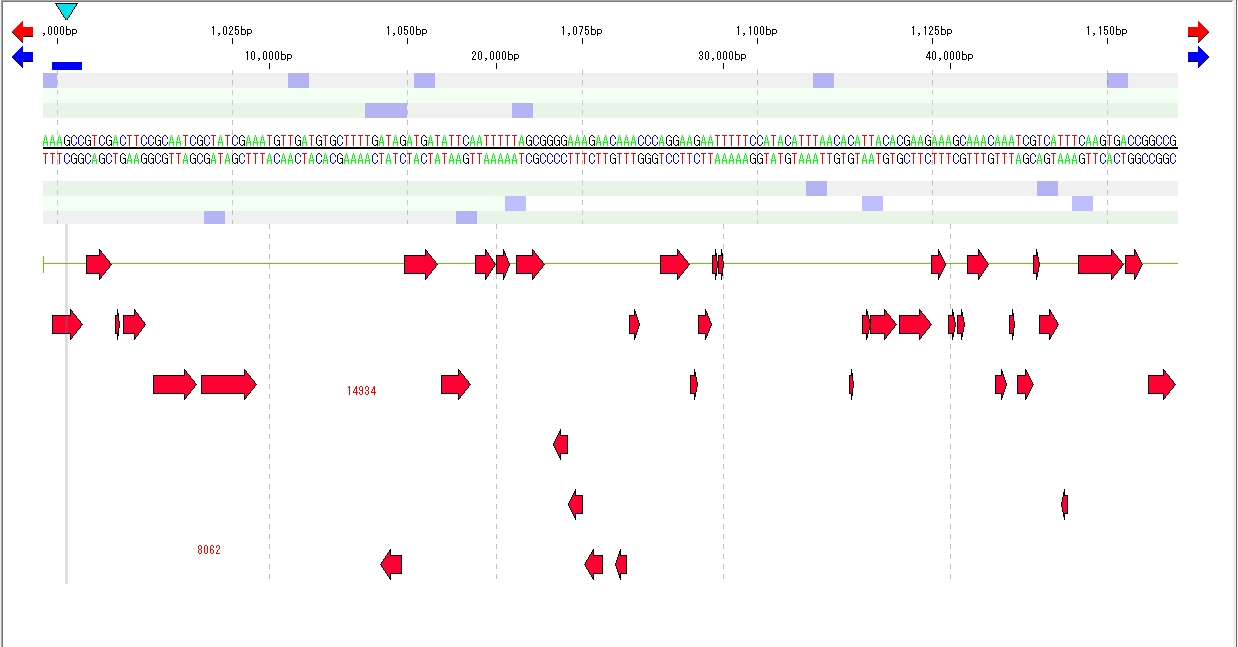
- The features registered in the main feature map are displayed.