Cloning
IMC is characterized by being able to perform operations of cloning experiment on computer. In this case, special data is unnecessary and it is possible to clone DNA sequence data which can be obtained from a public database such as GenBank or EMBL as it is. For cloning, restriction enzyme digestion, PCR primer design, PCR amplification, and ligation can be performed without changing the annotated sequence. All resulting cloning products are output in GenBank / EMBL format. Since Primer information is pasted and stored on the DNA sequence, it is also useful for Primer management.
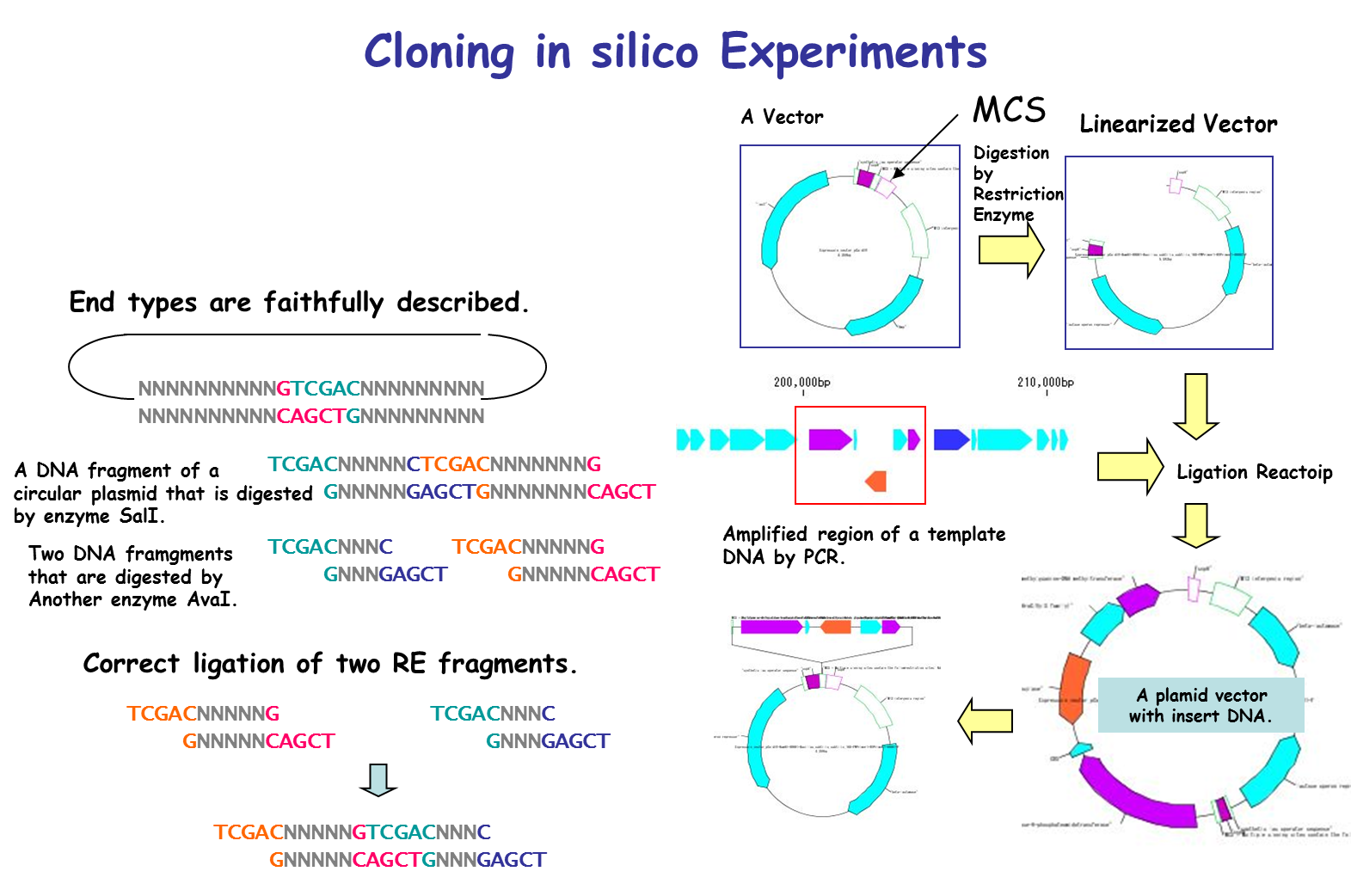
It is a cloning function that can actually cut / ligate.
in silico cloning experiment is possible.
It will help you to understand invisible molecular biology experiments. It can be used for assisting and simulating molecular biology experiments.
You can construct arbitrary Vector / Plasmid.
We will list optimal restriction enzymes for Vector insert check and simulate the gel electrophoresis results.
- Restriction enzyme treatment (restriction enzyme map, restriction enzyme digestion fragment generation, optimal restriction enzyme candidate list for insert check, others)
- PCR primer design,
- Primer design to avoid specific features,
- PCR replication (including annotation),
- Batch primer design to amplify all genes,
- Primer management ligation, self-ligation,
- Checking compatibility of base fragment end shape
- Plasmid map creation (Insert area blowing function) (Layout style correspondence)
- Addition of restriction enzyme recognition site to cloning DNA terminus with annotation described
- DNA terminal blunting, phosphorylation (dephosphorylation
- Arbitrary region extraction of annotated base sequence
 ドングルライセンス(HWキー)
ドングルライセンス(HWキー) フィーチャーマップとレーン
フィーチャーマップとレーン 配列・データ入出力
配列・データ入出力 GenBank EMBLビューワ
GenBank EMBLビューワ 配列ビューワ
配列ビューワ アノテーションビューア
アノテーションビューア 環状ゲノムマップビューワ
環状ゲノムマップビューワ プラスミドマップビューア
プラスミドマップビューア トレース波形ビューア
トレース波形ビューア 分子系統樹ビューア
分子系統樹ビューア フィーチャーキー検索
フィーチャーキー検索 パターン検索
パターン検索 バッチ(一括)ホモロジー検索
バッチ(一括)ホモロジー検索 プライミング部位検索
プライミング部位検索 制限酵素解析
制限酵素解析 プライマー設計
プライマー設計 コンティグブリッジ
コンティグブリッジ DNA断片修飾
DNA断片修飾 相同組換処理
相同組換処理 DNA組成解析
DNA組成解析 コドン解析
コドン解析 ORF解析
ORF解析 配列データベースの管理
配列データベースの管理 多重環状ゲノムマップ
多重環状ゲノムマップ ドットプロット解析
ドットプロット解析 ベンダイアグラム解析
ベンダイアグラム解析 逆相補鎖配列の生成
逆相補鎖配列の生成 設定・インストール
設定・インストール インシリコアセンブラー
インシリコアセンブラー アイ・スパイダー(iSpider)
アイ・スパイダー(iSpider) タクシースパイダー (TaxiSpider)
タクシースパイダー (TaxiSpider) ARM
ARM GenBank GBFF Expander
GenBank GBFF Expander GenBankファイルチェッカー
GenBankファイルチェッカー
