Expression Analysis
- High-precision tiling array data can be browsed and analyzed in conjunction with annotations of genes etc. Affymetrix, Agilent and Nimblegen arrays are supported.
- Array profile parallel display (corresponding to layout style)
- Array data correction, array information statistics (graph creation)
- Array-to-array operation, Save calculation result file
- Probe level - Gene level expression intensity conversion
- Clustering by expression intensity by genes (between genes)
- Profile peak detection
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Subcategories
Probe Design 0
Automatically design tiling array probes.
You can specify the probe base length.
You can specify the distance (base number) between the leading bases of each probe.
You can specify the strand to design the probe (Forward Strand, Reverse Strand, Both). Probes can be designed avoiding specified annotations (feature keys) such as rRNA.
If there is any annotation at the position where the probe was designed, it is possible to capture its contents.
An arbitrary initial letter (prefix) and a sequential number of the specified number of digits can be specified for the probe name.
Currently it is possible to output in three different formats.
- Agilent Simple Format: Agilent Simple Format is a comma-separated CSV format of 2 columns per row consisting of ProbeID and Sequence.
- Agilent Complete Format: The Agilent Complete Format is a comma separated CSV format of 7 columns per probe consisting of ProbeID, Sequence, TargetID, Accessionns, GeneSymbols, Description and ChromosomalLocation.
- IMC Standard CSV Format: IMC Starndard CSV Format is a Comma Separated Probe Description CSV Format of 5 columns per row of 1 probe standardly used by IMC.
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Probe Mapping 0
This is a function to map a probe to annotated nucleotide sequence file.
You can read the probe file and map it to the origin position on the current genome sequence.
When there is position information on the genome, paste the probe feature to the corresponding genome position based on the position information of each probe.
If position information does not exist and only the base sequence of the probe is given, mapping is performed based on the homology of the probe base sequence.
Currently mappable probes belong to the following array makers.
- Affymetrix
- Agilent
- Nimblegen
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Import Expression Data File 0
Import tiling array expression data files and NGS RNA-Seq files so that they can be used for array expression analysis.
There is no limit on the number of expression data files that can be imported, but you can only import 10 files with one operation.
Probe mapping must be performed before this function can be executed.
You can load a genome base sequence file, map a probe file, import a expression data file, register it as a batch process, and execute it in the background.
The expression level for each probe can be displayed as a bar graph or a line graph on the array profile lane of the main feature map.
There is no limit on the number of lanes to display at the same time, but as you increase the number of lanes, it consumes more memory.
Expression Profile Lane Operations 2
Any number of expression profile lanes can be displayed on the main feature map.
Also, you can move its position freely up and down.
Expression profiles belonging to that region are also inherited if genomic sequences with expression profiles are digested, truncated, amplified, ligated, etc. by cloning operation.
This function is installed in the following software edition
IMCAE![]() , IMCDS
, IMCDS![]() , GenomeTraveler
, GenomeTraveler![]()
On the expression profile lane you can perform the following operations.
- Setting lane style
- Import color data
- Clear color data
- Output CSV format file of expression profile
- Profile Magnified
Expression Data Correction 0
This function corrects the expression profile data.
Data correction functions include the following.
- MM Subtraction from PM
- Quality Trimming
- Saturation Trimming
- Global Rank Trimming
- Global Rate Trimming
- Base Window Rank Trimming
- Probe Window Rank Trimming
- Probe Window Rate Trimming
- Border Line Trimming
- Base Window Rate Trimming
- R-I Conversion
- Level Conversion
- Trimmed Mean
- Trimmed Median
These data corrections are pipelined to execute multiple correction items in an arbitrary order.
The applied state of these data corrections is shown on the list displayed for each expression data file.
A statistical number of correction values of the results of these data corrections can be displayed in a list for each expression data file.
The distribution of the expression levels before and after the data correction can be displayed in a graph.
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Gene Level Data Conversion 0
Lengths of probes of tiling arrays are usually shorter than genes, and multiple probes are designed at approximately equal intervals on genes and between genes.
In addition, the measured primary data is the expression level for each probe.
Therefore, focusing on one gene, multiple probes and multiple expression levels can be obtained there.
This function converts the expression level of the probe placed on the gene into the expression level of the gene.
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Inter-Array Analysis 0
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Peak Detection 0
Detect peaks from the expression profile.
Detection results are listed in the Peak List dialog.
The list can be saved as a CSV file.
When you click on the list, the main feature map will automatically scroll so that its position is centered.
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Correlation Plot between Arrays 0
Plot the correlation of expression levels between arbitrary arrays.
- X-Y plot
- R-I plot
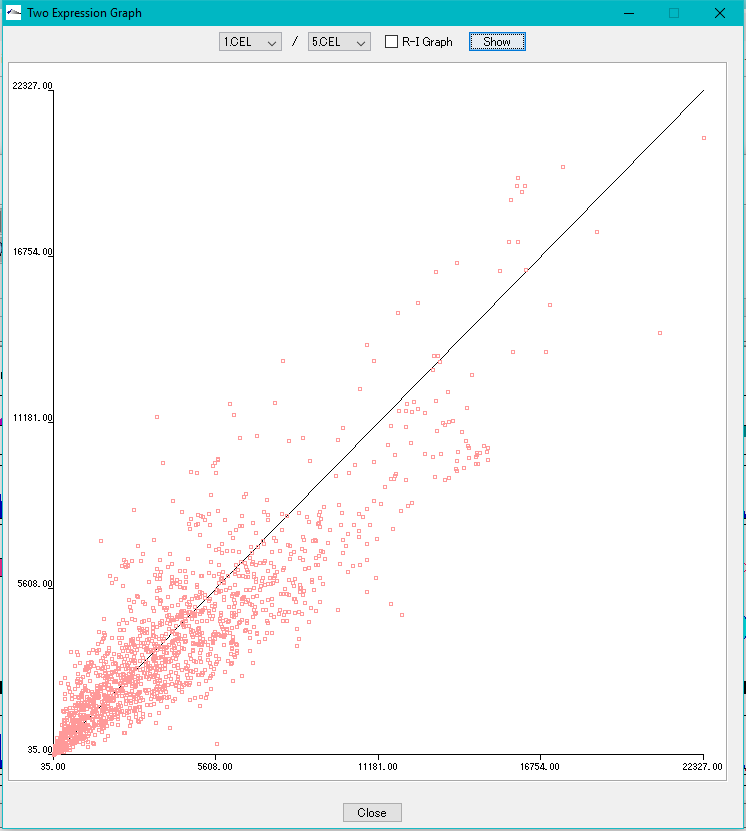
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Expression Clustering 0
Clustering of gene expression levels.
Clustering results can be displayed as dendrograms and heat maps.
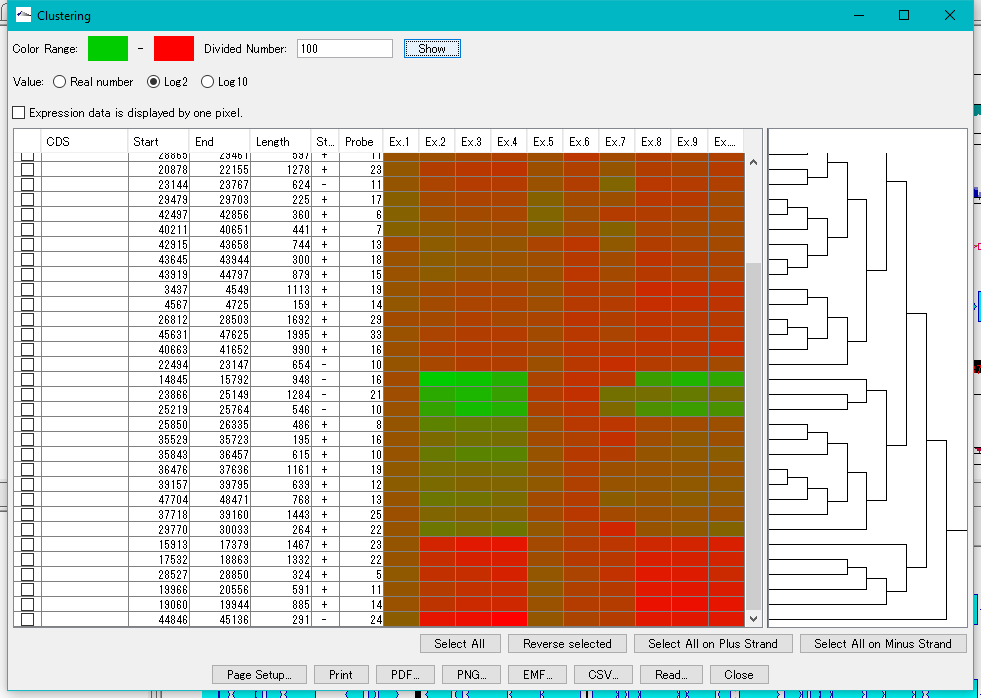
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Feature Expression Statistics 0
As for the presently imported expression level data file, the expression levels by feature are listed.
Features to display in the list dialog can be selected (multiple selections possible).
The list can be sorted with up to four sort keys per column.
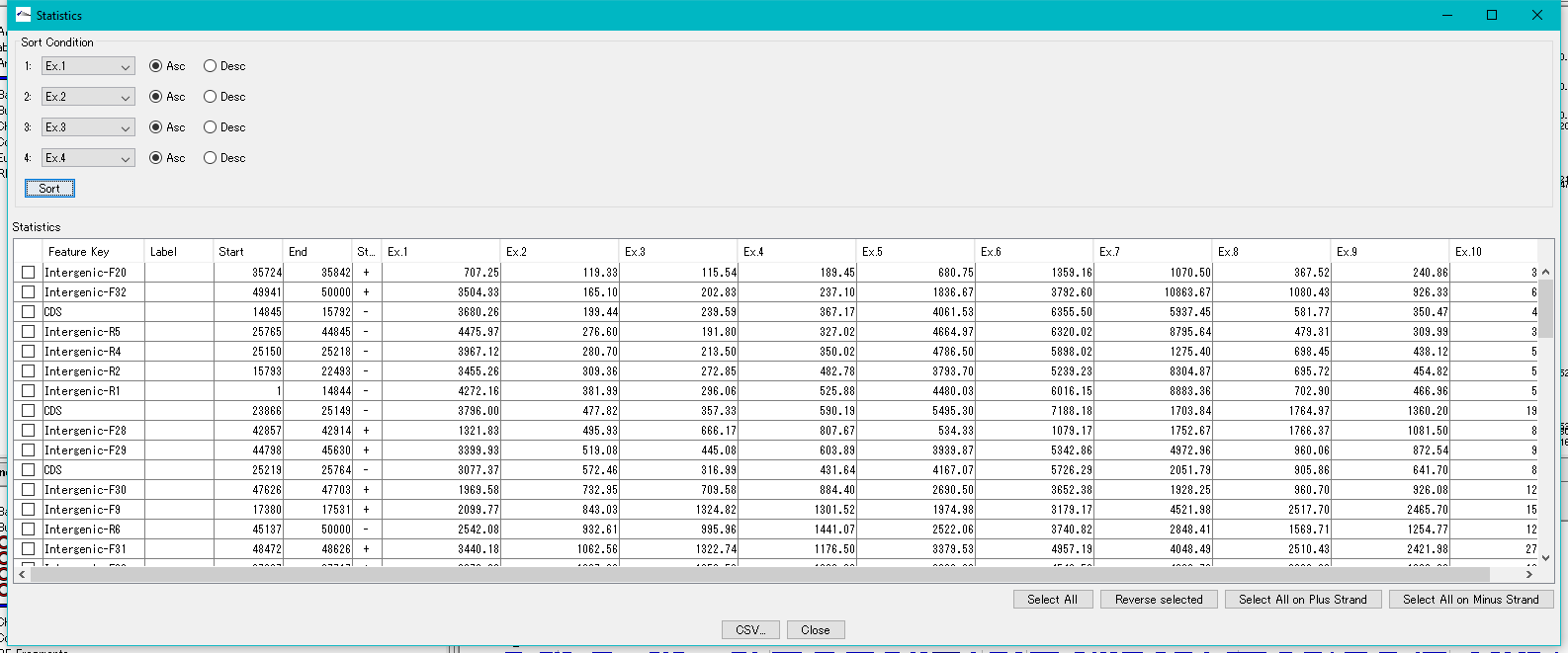
This function is implemented in the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
Arithmetic Operation between Arrays 0
Arithmetic operation between arrays is executed, and the result is displayed in the profile lane.
It is possible to calculate up to 3 arrays.
This function can be executed with the following edition.
AE![]() , DS
, DS![]() , GT
, GT![]()
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings