Feature Map
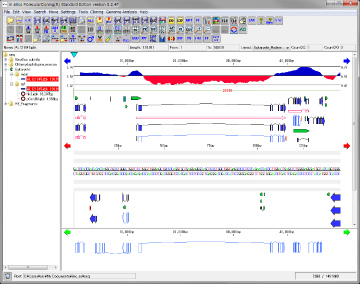 |
Genome feature map creation / drawing function
|
Subcategories
Feature Layout Style 23
By using the feature layout style, you can easily draw feature maps with complicated feature layouts by applying pre-registered layouts when drawing a feature map by combining multiple functional lanes.
Sequence Lane 9
In the sequence lane, the base of the currently loaded genome sequence file and the amino acid sequence if it is the coding region are displayed for each strand.
A sequence lane can be placed anywhere in the feature map. You can place more than one sequence lane on one feature map if necessary.
Four bases and 20 amino acids can be displayed individually in different colors.
Changing the drawing color and font size is done in the Sequence Lane tab pane of feature setting dialog.
You can change which position of amino acid 1 character code is placed in codon.
You can not change the font type.

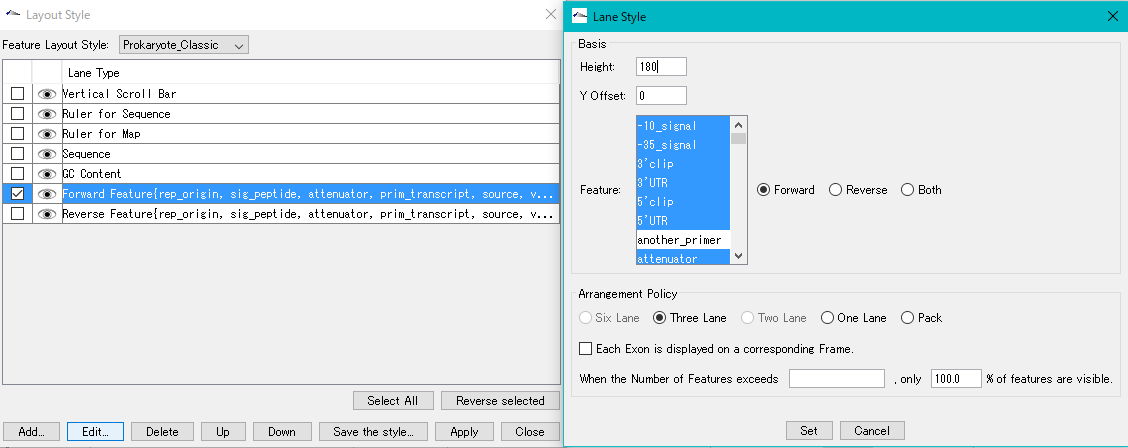
Feature Lane 12
- The above figure is a feature lane of the forward strand in three frame format.
- The following functions can be executed from the feature lane setting dialog.
- Display feature key (s), strand selection, arrangement method (Six Lane, Three Lane, Two Lane, One Lane, Pack), Chain change (Forward, Reverse, Both), Exon display method change, Placement density, Lane Height change, offset change.
Content Profile Lane 3
Content profile lane is a lane for displaying numerical values such as base composition, sliding window method and their profile.

The registered profiles that can be displayed in the content profile lane are as follows.
- GC Content
- GC Content
- AT Content
- A Content
- C Content
- G Content
- T Content
- GC Skew
- AT Skew
- Cumulative GC Skew
- Cumulative AT Skew
- Fickett
- Import Map Data
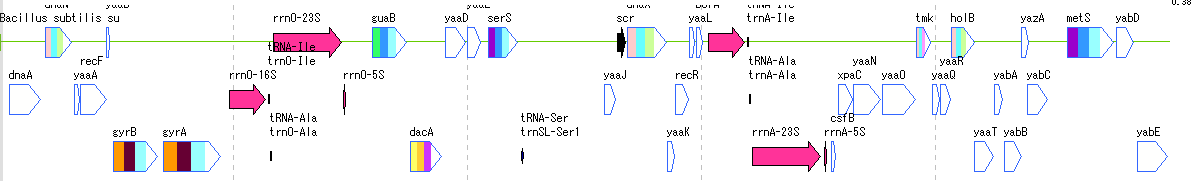
Navigation Lane 1
Navigation Lane displays the bird's-eye view map of the entire Feature Map, and you can move the display position of the Feature Map with the slider.
- The items that can be changed from the navigation lane setting dialog are as follows.
- Feature (s) can be selected. Strand selection (Forward, Reverse, Both), placement method selection (Six Lane, Three Lane, Two Lane, One Lane, Pack), thinning setting change.
Restriction Enzyme Lane 1
- The items that can be changed from the restriction enzyme lane setting dialog are as follows.
- Lane height, set switching, narrowing down of restriction enzyme list (recognition sequence length, palindromicity, terminal shape (Blunt, Sticky), DAM / DCM, number of cleavage sites of target sequence, selection of restriction enzyme.
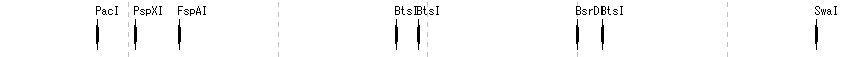
Frame Lane 3
- Frame Lane is a lane displaying areas where stop codons do not appear consecutively for each of six frames.
- For each of six frames, the region from the stop codon to the stop codon indicates the region with the specified number of codons or more.
- You can register as a new feature by right-clicking an area where stop codon does not exist.
LGRMap Lane 0
The local genome rearrangement map lane (LGRM lane) is a lane for drawing the local genome rearrangement map on the main feature map.
Display mutations in base units between two closely related strains.
The mutation list dialog is linked to this lane.
Implementation Edition
IMCGE![]() , IMCPE
, IMCPE![]()
Expression Profile Lane 0
Expression profile lane is a lane for graphically displaying expression data by tiling array or RNA-Seq.
Implementation Edition
IMCGE![]() , IMCPE
, IMCPE![]()
Multiple expression profile lanes can be registered and displayed in the main feature map, and the number of lanes that can be registered and displayed on one feature map is not limited.
When the number of display lanes increases, it is necessary to scroll vertically.

The lane can display the expression level depending on the genome position as a bar graph or a line graph.
You can change display and calculation settings for each lane.

Other Lanes 0
Vertical Scroll Bar Lane

Sequence Scale Lane and Map Scale Lane

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings