Tools
Subcategories
in silico Assembler 2
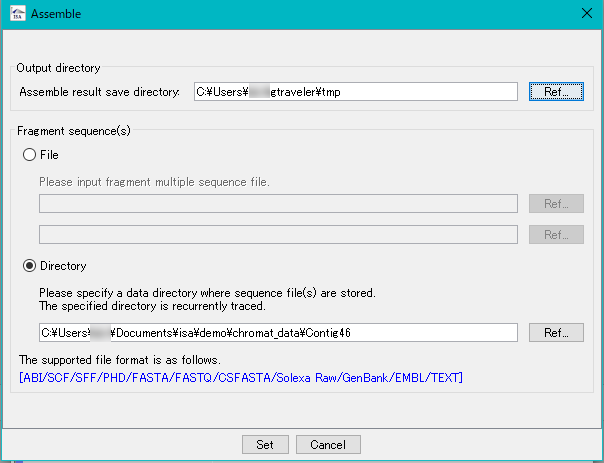
in silico Assembler is de novo Assembler.
You can assemble DNA fragments of 50 bp or more.
It is mainly used for assembly of fragment reads from capillary sequencer.
Reads from NGS can also be assembled (up to 1 million reads, please use Velvet etc for larger size).
In the preprocessing, limited number of processing, designation of minimum QV, maximum N bases can be limited.
iSpider 0
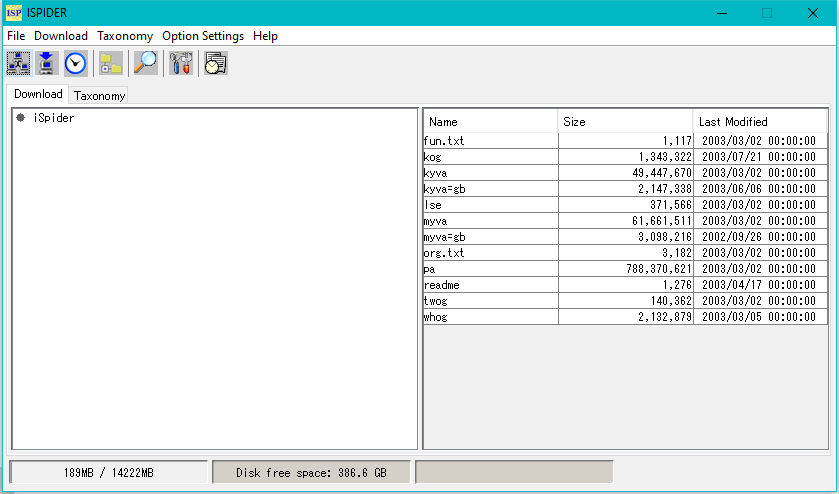
iSpider is a tool for downloading necessary sequence data files from various external sequence database servers.
By registering information of the external servers and the sequence data files acquired from it in advance, you can easily download the latest sequence data and save it in the specified local directory.
Although it is possible to execute the download immediately manually, it is also possible to specify the date and time to execute and download it automatically at that time.
Because it is possible to specify multiple files with regular expressions, in some cases you will download quite a lot of files.
Please do not load much load on the servers according to the rules of external server.
TaxiSpider 0
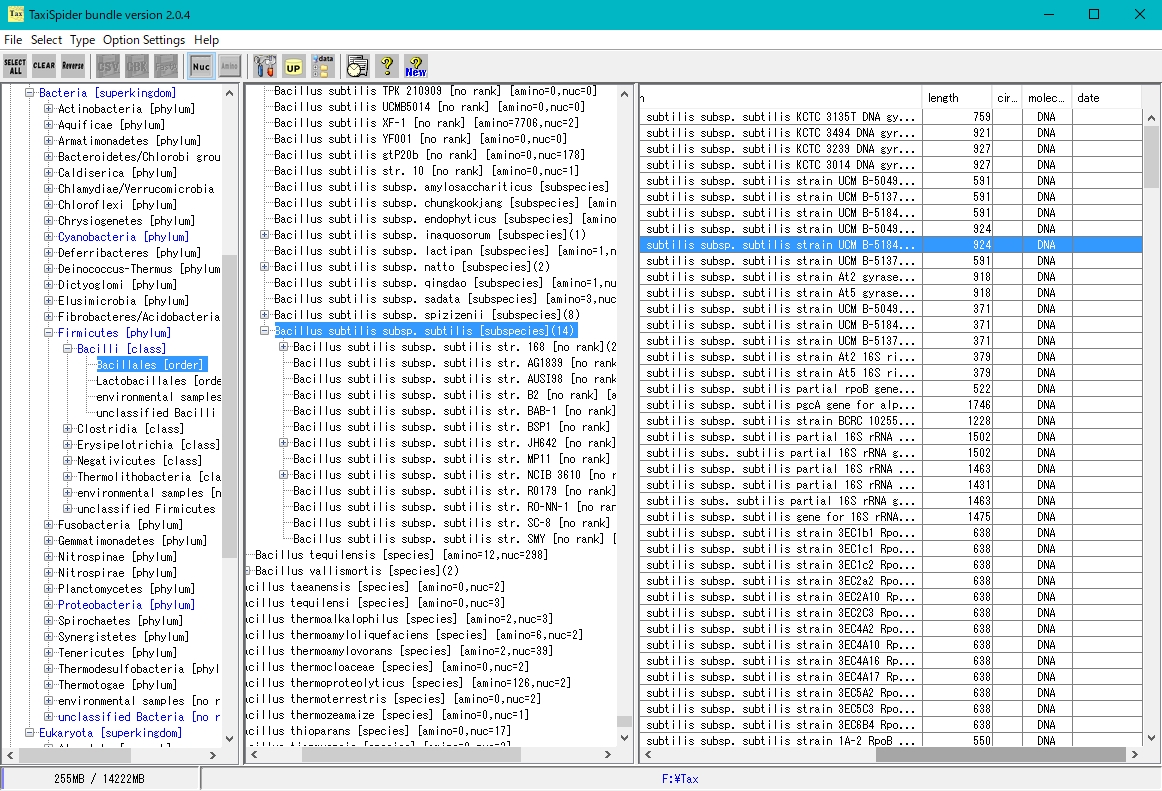
TaxiSpider is a sister tool of iSpider.
It is a tool to download all the indexes to the base sequence file and the amino acid sequence file from the specified site, but it is characterized by storing those array indexes in one big Taxonomy Tree like database.
The downloaded array data is stored on the corresponding branch of Taxonomy Tree according to its Taxonomy information.
However, it requires very large disk space locally.
As of December 2018, 47 GB of space is required.
ARM 0
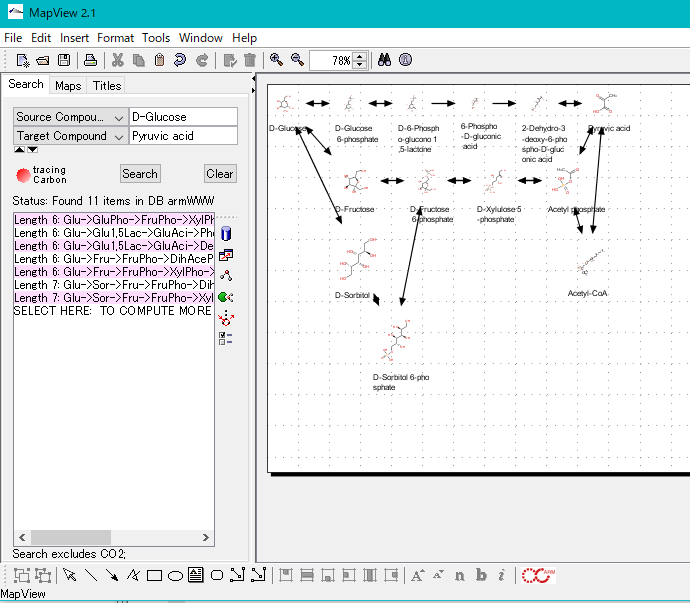
ARM is a metabolic pathway editor / viewer.
There is an atom trace function, which can track carbon, oxygen and sulfur atoms.
You can search all single pathways from source compound to target compound.
However, since the compound and reaction data are old, the latest pathway is not included.
It is possible to add compounds and reactions.
You can combine the searched pathways automatically on the canvas and draw a synthetic pathway.
The following new functions are also added to the original function.
You can import quantitative data for each compound obtained from metabolome analysis, etc., and display the series on each compound (bar graph).
In addition, expression level data of enzyme genes obtained from transcriptome analysis can be displayed in series (bar graph) on each reaction.
ARM is software developed by Prof. Arita of the University of Tokyo (present National Institute of Genetics).
Interactive ARM 0
It is an interactive ARM interface.
You can run ARM's single pathway discovery function interactively.
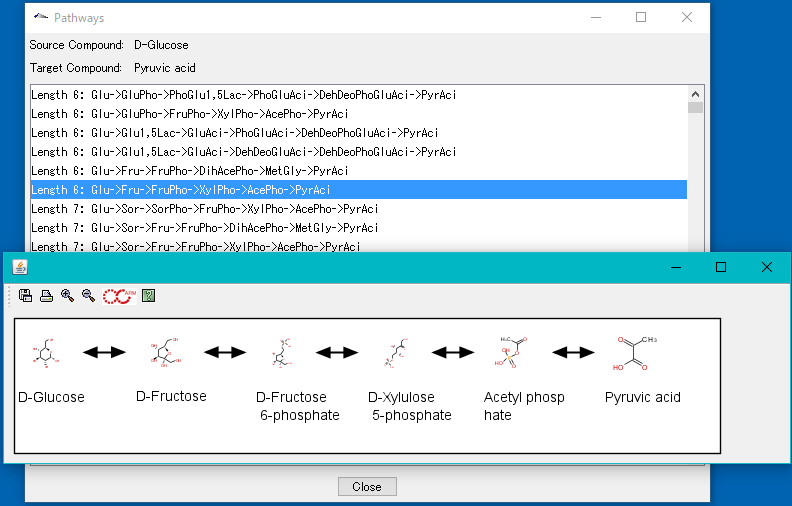
ARM is software developed by Prof. Arita of the University of Tokyo (present National Institute of Genetics).
Pair2Fragment 0
We convert NGS 's paired end read into fragments.
GenBank GBFF Expander 2
GenBank GBFF Expander is a tool to expand NCBI's GBFF format file containing a large number of genome sequences into individual genomic sequences and place them in the tree directory according to the Taxomy.
It is a prototype of TaxiSpider.
GenBank File Checker 0
This function checks whether the GenBank EMBL format file, which is the main format of the genome base sequence file used in IMC, is correct, and corrects it automatically if there are errors.
Storing multiple GenBank EMBL files under one directory will check all the files there.
In addition, the inspection result file can be saved in a different directory.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings