Cloning
IMC is characterized by being able to perform operations of cloning experiment on computer. In this case, special data is unnecessary and it is possible to clone DNA sequence data which can be obtained from a public database such as GenBank or EMBL as it is. For cloning, restriction enzyme digestion, PCR primer design, PCR amplification, and ligation can be performed without changing the annotated sequence. All resulting cloning products are output in GenBank / EMBL format. Since Primer information is pasted and stored on the DNA sequence, it is also useful for Primer management.
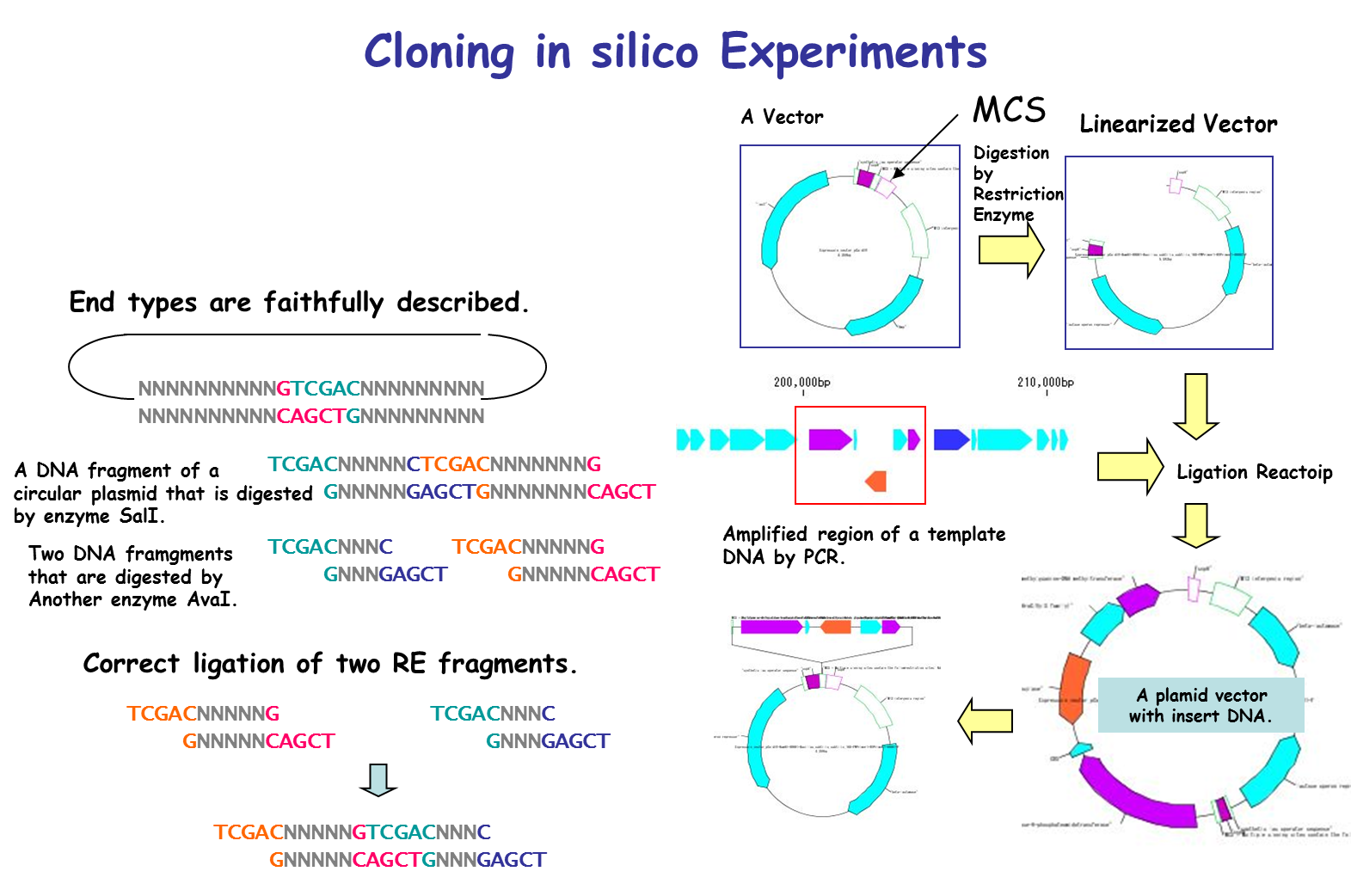
It is a cloning function that can actually cut / ligate.
in silico cloning experiment is possible.
It will help you to understand invisible molecular biology experiments. It can be used for assisting and simulating molecular biology experiments.
You can construct arbitrary Vector / Plasmid.
We will list optimal restriction enzymes for Vector insert check and simulate the gel electrophoresis results.
- Restriction enzyme treatment (restriction enzyme map, restriction enzyme digestion fragment generation, optimal restriction enzyme candidate list for insert check, others)
- PCR primer design,
- Primer design to avoid specific features,
- PCR replication (including annotation),
- Batch primer design to amplify all genes,
- Primer management ligation, self-ligation,
- Checking compatibility of base fragment end shape
- Plasmid map creation (Insert area blowing function) (Layout style correspondence)
- Addition of restriction enzyme recognition site to cloning DNA terminus with annotation described
- DNA terminal blunting, phosphorylation (dephosphorylation
- Arbitrary region extraction of annotated base sequence
Subcategories
Cloning in silico 1
Using IMC's in silico cloning function, continuous cloning experiment on PC or Mac is possible.
Digestion of the genomic DNA sequence with restriction enzyme, gel electrophoresis of the fragment, ligation to the opened vector, etc. can be carried out in succession.
Restriction Enzyme 12
With the restriction enzyme function of IMC, if annotation (Feature) is added to the original DNA base sequence, it can be fragmented with the annotation kept.
If one feature is divided by a restriction enzyme, its feature itself is inherited, but in some circumstances the feature key may be changed automatically.
The terminal shape of restriction enzyme digested fragments is preserved correctly. By doing this, we check whether ligation is possible when ligation between pieces is done.
In E. coli, it can be checked whether the methylation site affects restriction enzyme digestion.
It is also possible to register frequently used restriction enzymes as a set.
It is also possible to register newly provided restriction enzymes or delete restriction enzymes that are no longer used.
Restriction enzyme management
- Partial set selection,
- Filtering by type
- Distinction of the number of recognized bases
- Palindromic
- Distinction between smooth and protruding ends
- Presence / absence of DAM / DCM
- Difference depending on the number of cutting points
- Alias names of restriction enzymes list can be saved, list editing is possible.
- You can also register enzymes, edit enzymes, delete enzymes.
Search function of restriction enzyme recognition site
- Search area selection,
- Recognition site search,
- Recognition site list display,
- Restriction map display,
- Collective search from multiple sequences
Primer Design 15
IMC has various PCR Primer design methods.
The ability to simply drag the nucleotide sequence on the sequence lane and register it as a primer
- Register primers from the sequence lane
- Register primers from the feature lane
Ability to design features on the feature lane and primers to amplify inside and outside the selected area
- Primer design for amplifying included products, including selected features of feature lanes
- Primer design for amplifying the product containing or containing the selected genomic region of the feature lane
A function of designing primers that amplify a large number of areas at once (with Iterate Design function: it is a function to repeat the design until there is no area that can not be designed)
- Batch PCR Primer Design Batch PCR primer design to amplify all features with specified feature key on genome
- Whole Genome Covering PCR Primer Design PCR primer design that PCR product completely overlaps whole genome or large genomic region with PCR product overlapping
- Sequencing Primer Design: Sequence primer design for sequencing DNA fragments that can not be covered with one lead of a capillary sequencer
reference
The following is a function to design a group of primers for cloning multiple DNA fragments at once, such as gene cluster design. From design to cloning, you can load cloned products into the IMC.
- In-Fusion Design Ver.1: Primer design function for In-Fusion Cloning
- In-Fusion Design & Build Ver.2: Primer design function for In-Fusion Cloning
- In-Fusion Design & Build Ver. 3: Ability to design and build gene clusters by dropping DNA fragments to be inserted into vector sequences
- Batch Gene Cluster Design & Build: the ability to design and build lumped numerous gene clusters by replacing the gene sets that make up the cluster
- Combinatorial Design & Build: The ability to combinatorially combine design and build a fragment constituting a gene cluster
PCR Reaction 3
In the PCR function of IMC, the priming site of the registered primer set is searched from the genome base sequence. If both primers have a priming site on the genome, PCR amplify the region sandwiched between the primers.
In this case, adenine can be made to protrude by 1 base. It is used for TA cloning.
An annotation is inherited also in the amplified PCR product when the template DNA sequence is annotated (Feature).
It is also possible to perform PCR on multiple template DNA sequences.
Ligation 3
Purpose and overview
- Ligation is an essential experiment to perform molecular cloning. IMC combines various nucleotide sequences including annotated nucleotide sequences with each other in the same manner as in actual cloning experiments to generate the products. All annotations are properly succeeded also to the ligation products and their products can be analyzed using many functions of the IMC as well as the original base sequence.
Function
- If the terminal sequences complementarily match each other, ligate them into one base sequence.
- Up to 5 fragment segments can be ligated.
- You can generate all possible ligation sequences.
- In Version 7.24 and later, the complete complementary sequence of the ligation product is counted as a separate product.
- Annotated nucleotide sequences of GenBank and EMBL format can be ligated as well, and these annotations are properly inherited by the ligation products.
- If the shapes and sequences of the ends of the base sequence complementarily match, a circular base sequence is generated.
- It displays a list of restriction enzymes that can be used to check the insert direction of each fragment in the generated ligation product.
- Also displays gel electrophoresis pattern when using these restriction enzymes.
- Displays the terminal shape of all base sequences loaded in the current folder.
- Self-ligation can also generate circular DNA.
- It also supports TA cloning of PCR products.
- Before ligation, you can see the end shape of each piece.
- Plasmid map of the ligation product can be drawn. In this drawing, you can represent the insert area with a balloon
- The result of binding by ligation is a covalent bond (phosphodiester bond) equal to the bond between other bases, so after binding it can not distinguish originally.
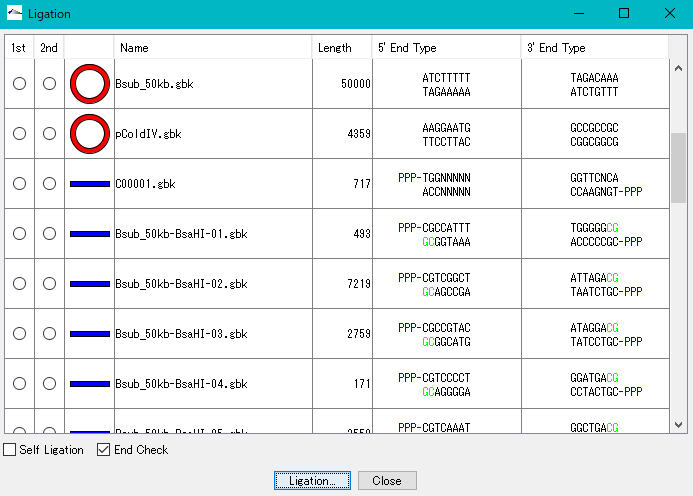
- The base sequence generated as a ligation product can be analyzed in the same way as the original base sequence.
Restrictions
- Currently, ligation linking 6 or more base sequences is not possible.
Algorithm
- For restriction enzyme cleaved sequences on IMC, special Feature Keys and Qualifiers are generated that indicate the terminal status.
Bridge Contigs 0
When a sequence of PCR products is searched for a group of contig sequences and the PCR product bridges between two contigs, combine the contigs into one sequence.
Plasmid Map 1
For Plasmid Map creation, please see Plasmid Map Viewer.
Gel Electrophoresis 1
When DNA sequence is digested and fragmented with restriction enzyme, when PCR amplification is carried out, ligation can be displayed as results of gel electrophoresis of those products.

Fragment End Modification 0
You can modify the ends of DNA fragment sequences.
The terminal modification functions currently provided by IMC are as follows.
Molecular biological treatment
- Blunting (blunting by T4 DNA Polymerase, blunting by Mung Bean Nuclease)
- Phosphorylation
- Dephosphorylation
- Adenine overhang on PCR product
Non molecular biological treatment
- Production of sequence with protruding thymine (T-Vector)
- Attaching of restriction enzyme recognition sequence
Find RE Set for Insert Check 0
If both ends of the DNA fragment to be inserted are cleaved with the same restriction enzymes or have blunt ends by ligation etc., the optimal restriction enzyme candidate for displaying the orientation of the insert is indicated by gel electrophoresis diagram .
Homologous Recombination 0
In the genome design function, a homology arm for homologous recombination is designed from the region to be homologously recombined and the insert sequence.
This function is used to recombine the insert sequence with the designed homology arm into the genome.
Pseudo Cloning 0
Regardless of molecular biology, base sequence processing by information manipulation
- Delete Sequence: Deletes an arbitrary region of the genomic base sequence.
- Fragmentation: Cut the genomic base sequence at the specified base position (multiple designation possible) and fragmentes it.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings