Gene and Genome Sequence Analysis
Explanation of basic composition analysis of genes and genomes, codon analysis, ORF extraction, amino acid translation, amino acid profile analysis, motif analysis and so on.
Subcategories
DNA Content Analysis 1
IMC analyzes the base composition of the genome sequence.
You can perform this analysis on sequence files loaded in the current sequence directory.
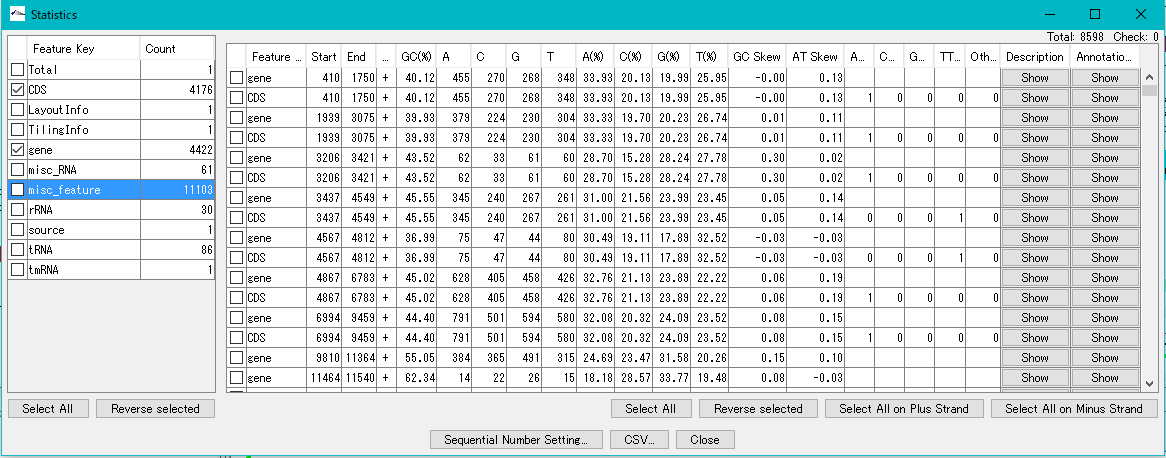
The Feature Statistics function of the Genome Analysis menu outputs the base composition of all the features in the currently loaded nucleotide sequence file.
The content profile lane that can be displayed on the main feature map is also one of the functions of displaying base composition.
Changes in composition by moving average method are displayed graphically.
The content profile lane is also implemented in the circular genome map viewer.
The "Cluster Design Checker" function evaluates the base composition of the specified cluster and checks whether it is in the preset base composition range or not.
Codon Analysis 0
IMC outputs the codon composition and amino acid composition table of coding region (CDS).
You can perform this analysis on annotated sequence files loaded into the current sequence directory.
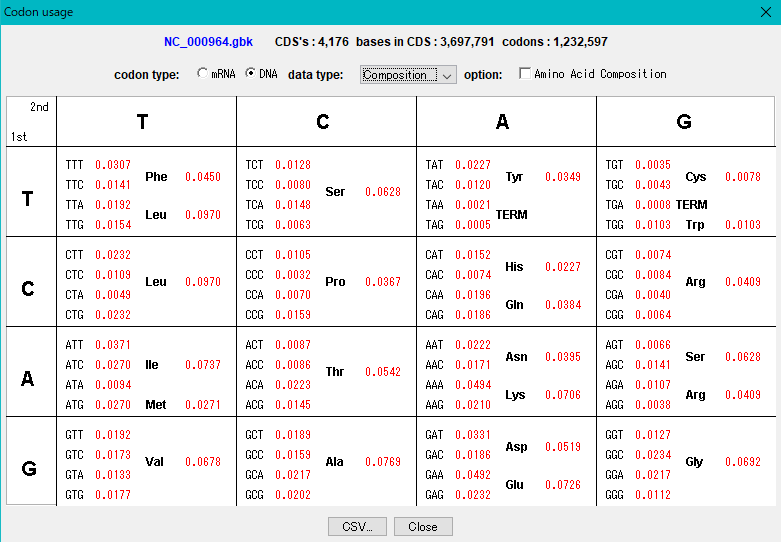
The "Show Codon Usage ..." function in the Genome Analysis menu not only outputs the individual amino acid composition and codon composition of all the CDSs identified in the currently loaded sequence, but also the amino acid composition of the total CDS , Codon composition can also be output.
Even with the Statistics function, the type of start codon is output for each CDS.
The codon substitution function "Change Codon" allows you to replace the codon composition according to the specified Codon Usage file.
Open Reading Frame Analysis 4
IMC provides a simple ORF candidate extraction function.
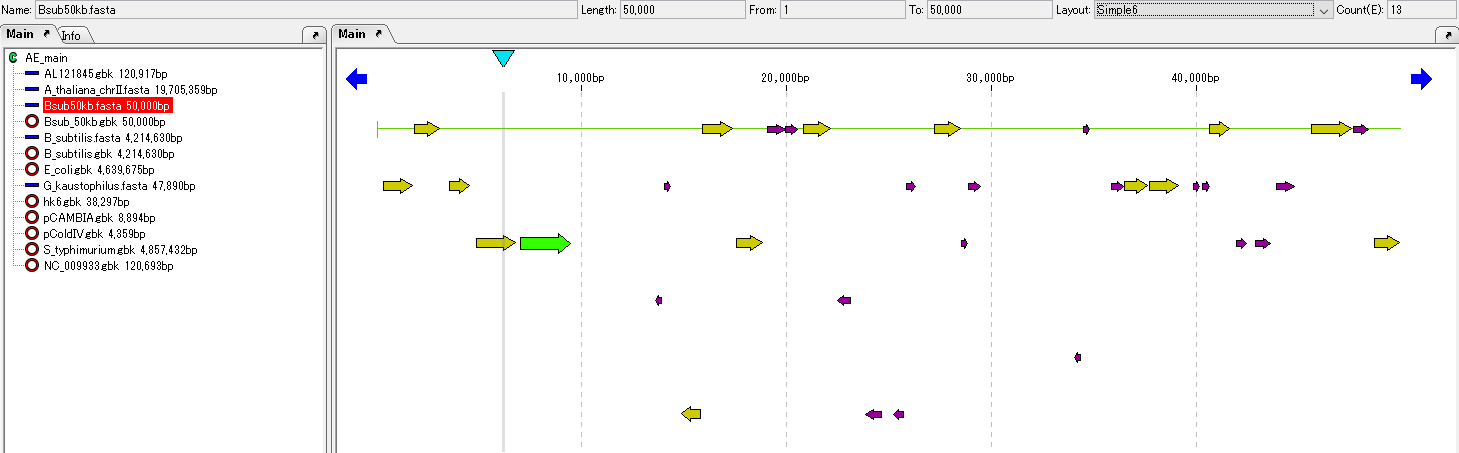
Extracts on the 6 frames of the current base sequence those having a length equal to or longer than the base length specified in the region from the stop codon to the next stop codon.
ORF candidates can be converted to CDS and amino acid translation can also be performed.
When the ORF candidate region encompasses other ORF candidate regions, one with a larger base length can be adopted.
In IMC, in addition to this, you can launch the Gene Finding programs Augustus and MetaGenome Annotator as external commands and capture the results.
In addition, you can import the output result of a gene identification program such as Glimmer and map it on the current sequence.
Translation 2
The base sequence of the CDS feature on the current base sequence is translated into amino acids.
You can specify the translation range for all CDS on current sequence, CDS on selected area, and one CDS.
Each codon is translated according to the specified Genetic Table.
Amino Acid Profile Analysis 0
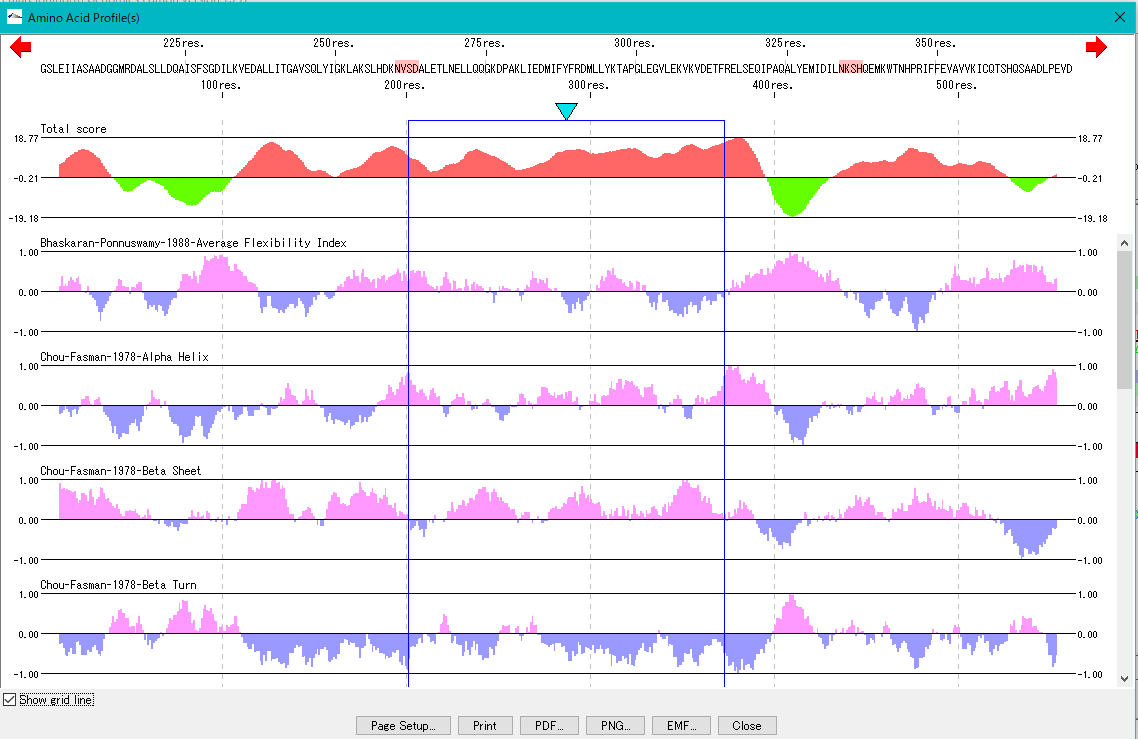
The secondary structure of the amino acid sequence of the gene are shown as a profile.
Structures such as alpha helix, beta sheet, turn, etc., profiles such as hydrophilicity index, hydrophobicity index, surface quality, chain flexibility, etc. can be displayed in parallel.
Also, you can freely create a comprehensive index that linearly combines these indicators.
Amino Acid Motif Analysis 0
If a motif is registered in the motif list of amino acid profile analysis, the position of the motif can be displayed on the amino acid sequence of the amino acid profile display dialog.
Amino acid motif search can be executed from pattern search function.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings