Function Overviews and Basic Operation
|
|
Subcategories
Launch 10
There are the following methods to launch IMC.
- Double click the desktop icon
- Launch from the start menu (Windows)
- Launch from the command prompt (Windows)
- Launch from Terminal (Mac)
Launch IMC 7
There are the following methods to launch IMC.
- Double click the desktop icon
- Launch from the start menu (Windows)
- Launch from the command prompt (Windows)
- Launch from Terminal (Mac)
Launch GT 2
There are the following methods to launch GT.
Double click the desktop icon
Launch from the start menu (Windows)
Launch from the command prompt (Windows)
Launch from Terminal (Mac)
Terminate 5
There are the following ways to terminate IMC.
Normal Termination of IMC
- Select the Exit menu and exit
- Click the "X" button in the main window to exit
- In the case of the above normal termination, the changed various setting parameters are automatically saved.
- Sequences that were being edited in the current sequence directory may be saved automatically, and prompts to save or not may be displayed. You can select this setting in the Feature Setting -> Setup tab pane.
Forced Termination
- Forced termination of IMC (Windows)
- Forced termination of IMC (Mac)
- When forcibly terminating the IMC, the sequences that were in the current sequence directory just before may not be saved. Also, various changed setting parameters may not be saved.
Basic Application Operation 6
The following operations are often used in IMC.
- Load (read) the base sequence file and display the main feature map.
- Perform Keyword Search.
- Find the restriction enzyme recognition site.
- View the sequence from the reverse complement side.
- Perform homology search.
Use IMC for the First Time 0
Please visit when using IMC for the first time.
Use GT for the First Time 5
Please read the following when using GenomeTraveler (GT) for the first time.
Sample data will be installed automatically when GT is started for the first time.
Using this sample data, you can practice the operation.
If sample data is not displayed, confirm the demo directory and change it to the root directory and it will be displayed.
Sample data includes the following.
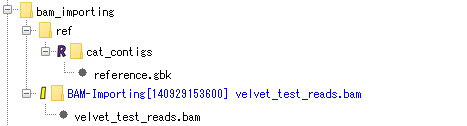
BAM file import:
Contains the result of importing the BAM file used for assembly and mapping. Each fragment of the BAM file can be viewed with the Alignment Viewer.
Mapping (using LAST):
The reference genomic data and the result of mapping NGS lead using LAST on its genome are stored. You can browse and analyze the reference genome data by activating the IMC, and you can view and analyze the mapping by starting the Alignment Viewer. It is also possible to execute new mapping using the same NGS data.

Assembly (using OASES):
Contains the AFG file of assembly result of NGS lead using OASES. Convert this to a BAM file and start the Alignment Viewer. It is also possible to use the same lead and execute a new assembly.

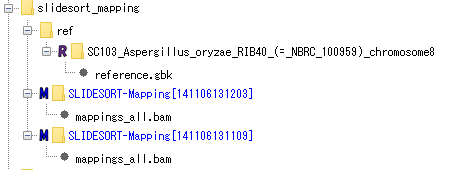
Mapping (using SLIDESORT):
contains reference genomic data and results mapped using SLIDESORT on its genome. You can browse and analyze the reference genome data by activating the IMC, convert the mapping result to a BAM file, and view and analyze it with the Alignment Viewer. It is also possible to execute new mapping using the same NGS data.
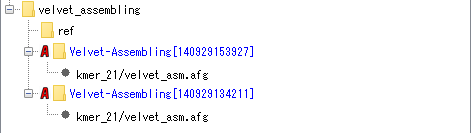
Assembly (using Velvet):
The AFG file of assembly result of NGS lead is stored using Velvet assembler. Convert it to BAM file and start up Alignment Viewer. It is also possible to use the same lead and execute a new assembly.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings