DNA Content Analysis
IMC analyzes the base composition of the genome sequence.
You can perform this analysis on sequence files loaded in the current sequence directory.
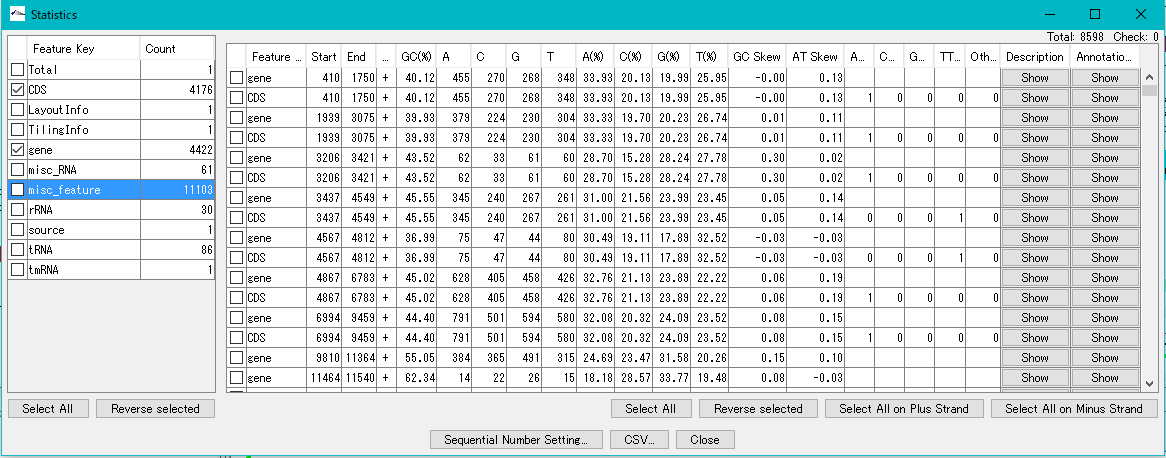
The Feature Statistics function of the Genome Analysis menu outputs the base composition of all the features in the currently loaded nucleotide sequence file.
The content profile lane that can be displayed on the main feature map is also one of the functions of displaying base composition.
Changes in composition by moving average method are displayed graphically.
The content profile lane is also implemented in the circular genome map viewer.
The "Cluster Design Checker" function evaluates the base composition of the specified cluster and checks whether it is in the preset base composition range or not.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings