ARM
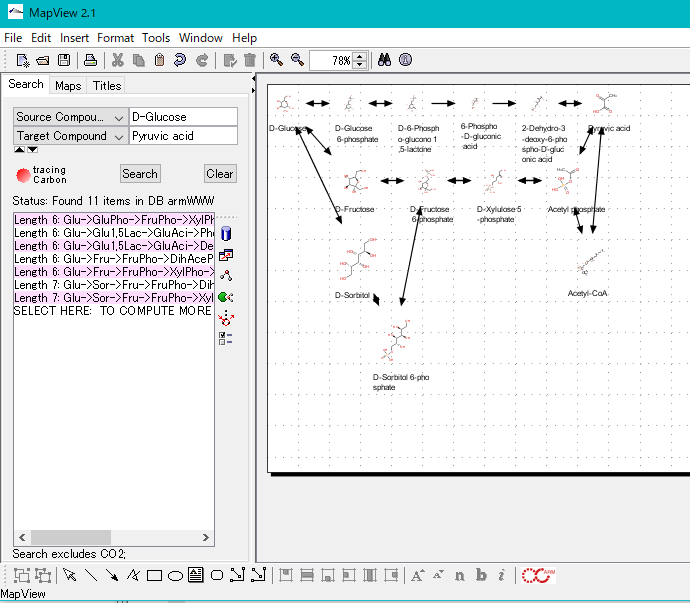
ARM is a metabolic pathway editor / viewer.
There is an atom trace function, which can track carbon, oxygen and sulfur atoms.
You can search all single pathways from source compound to target compound.
However, since the compound and reaction data are old, the latest pathway is not included.
It is possible to add compounds and reactions.
You can combine the searched pathways automatically on the canvas and draw a synthetic pathway.
The following new functions are also added to the original function.
You can import quantitative data for each compound obtained from metabolome analysis, etc., and display the series on each compound (bar graph).
In addition, expression level data of enzyme genes obtained from transcriptome analysis can be displayed in series (bar graph) on each reaction.
ARM is software developed by Prof. Arita of the University of Tokyo (present National Institute of Genetics).
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings