IMC F04G Functions Executed from Linear Genome Map
The following functions can be executed from the multiple linear genome map.
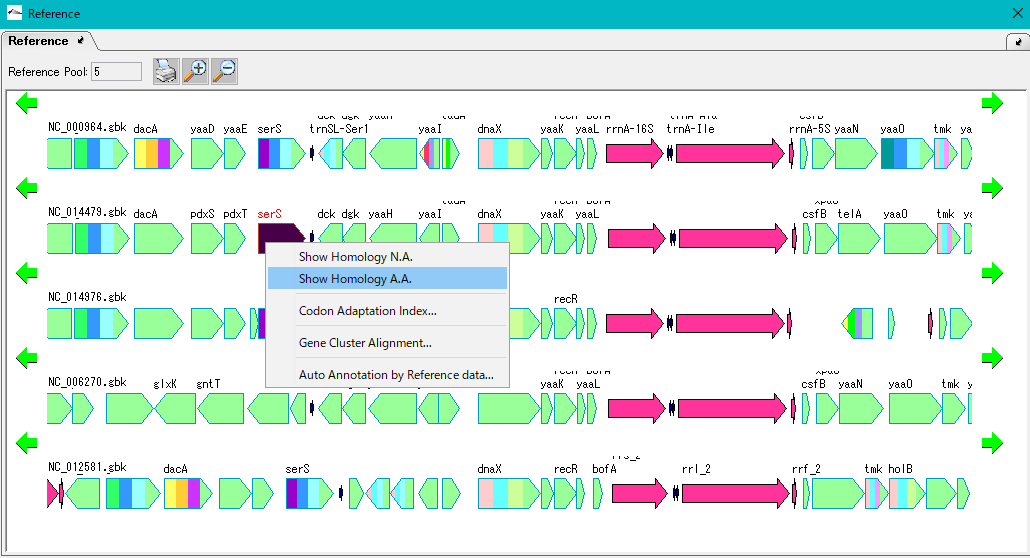
- Homology search (nucleic acid, amino acid)
- A homology search is performed on one CDS on the linear genome map against the other genome of the linear genome map and the CDS of the current sequence of the main feature map and as a result the genomic alignment on that CDS in the multiple linear genome map to hold.
- Gene composition change function for recombination into heterologous host
- Replace one CDS on the linear genome map with homologous genes in the current genome of the main feature map. At this time, we will automatically change the composition of recombinant genes based on Codon Adoptation Index.
- Gene cluster alignment
- With respect to one CDS on the linear genome map, cluster alignment is performed by performing gene cluster comparison with other genomes on the multiple linear genome map, including upstream and downstream CDS.
- Automatic annotation
- Run the automatic annotation on the current genome of the main feature map with reference to the linear genome map as the genome.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings