IMC S01A Browse Genomic Sequence from Reverse Complementary Strand
When IMC loads a genome sequence file into the main feature map, it normally displays the base sequence described in the file as a forward strand.
When annotations are described, the position of each annotation is also the base sequence position on the forward strand.
In IMC, there is a function called Reverse Comment. When this function is executed, it can be changed from the 5 'side of the complementary strand of the currently loaded genomic sequence to the forward strand and displayed.
At this time, change the annotation position from the original forward strand to the position on the current forward strand (original reverse complementary strand).
If you execute the Reverse Complement operation again, it will return.
When the reverse complementary strand is displayed, if the current genome sequence is saved, the stored nucleotide sequence is rewritten from the 5 'side of the reverse complementary strand of the original genomic sequence file b.
All annotations are also saved with the position rewritten.
Operation
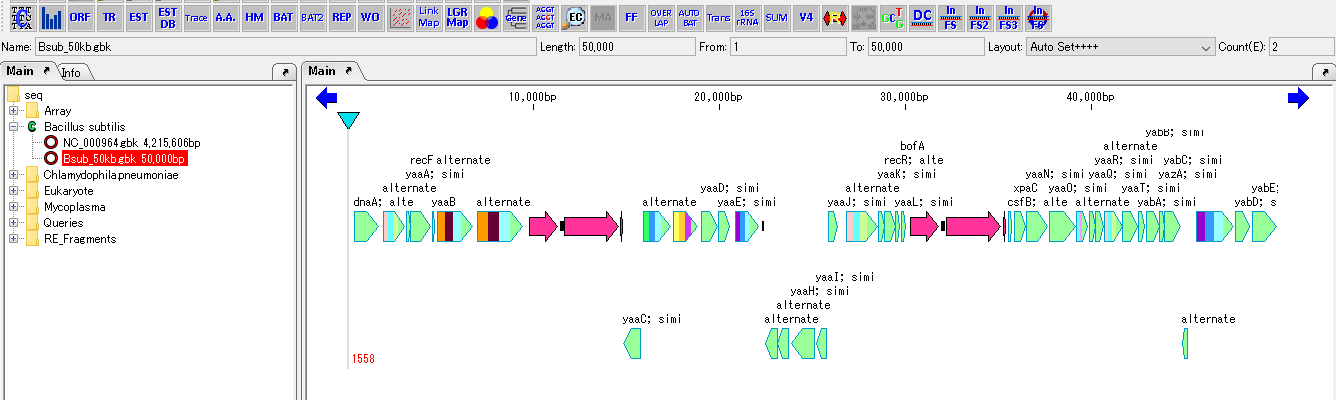
- Load the genome base sequence into the main feature map.
- Here, we will use the sample data Bsub_50kb.gbk.
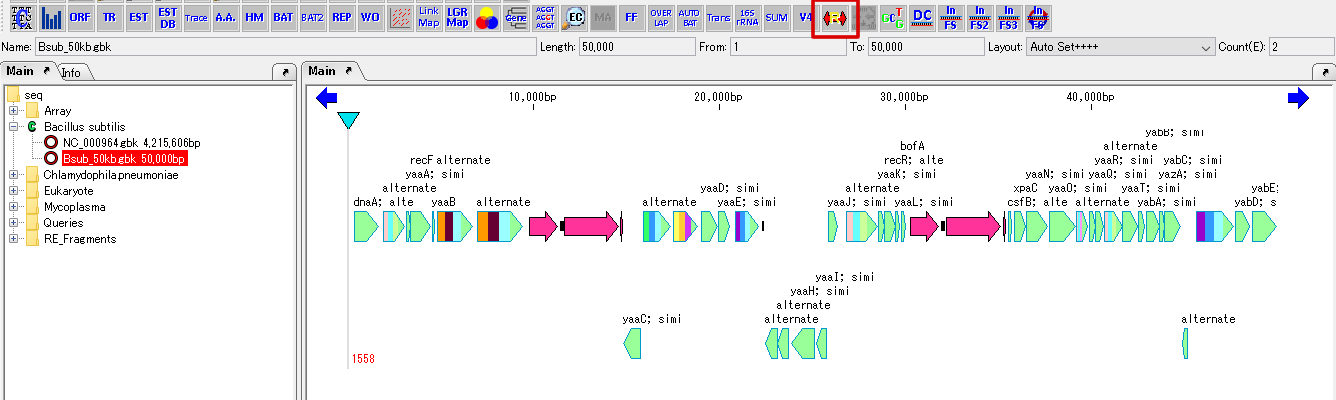
- The feature map of the loaded genomic sequence is displayed on the feature lane of the main feature map.
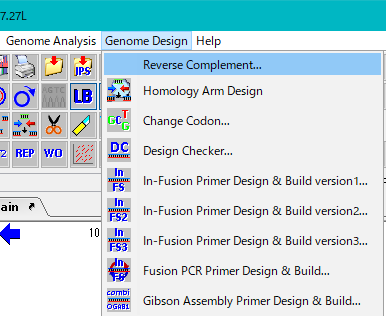
- From the menu click Genome Design -> Reverse Complement.
- Alternatively, click the Reverse Complement button in the toolbox.
- The forward and reverse complementary strands of the feature map are switched, and all the features also move and invert the display with the position on the original reverse complementary chain as the position on the ordinal chain.
- The color of the Reverse Complement button in the toolbox also changes, indicating that the reverse complementary side is currently displayed.
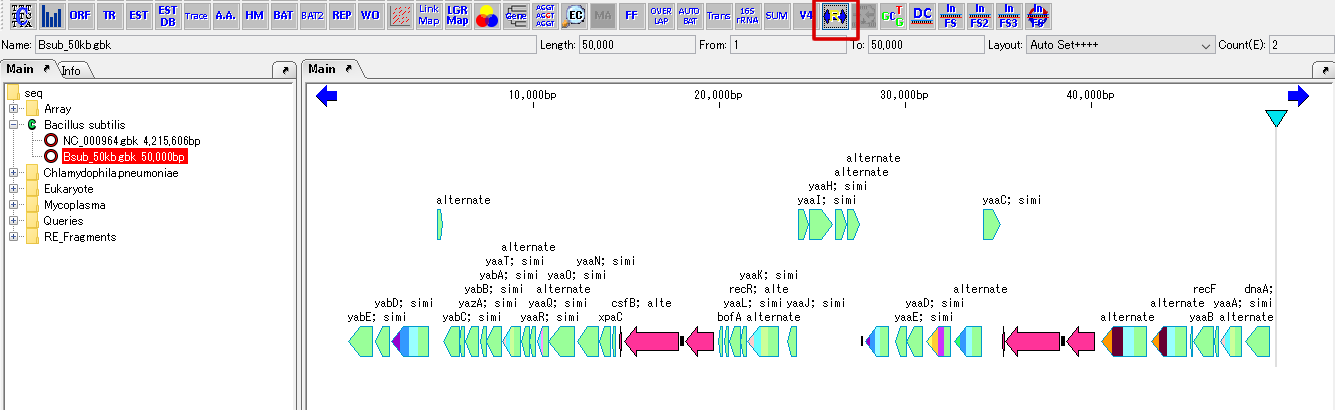
- The position and direction of the feature have been changed internally, but the original array file has not been replaced.
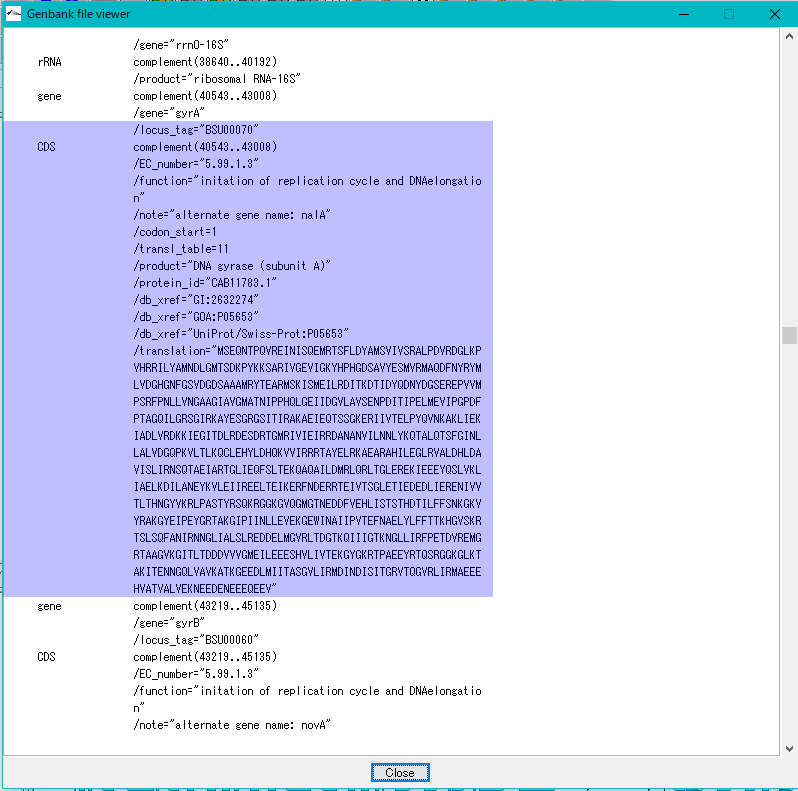
- If you click on one feature here and then click on GenBank / EMBL Viewer and browse the description of that feature, that position still shows the original position.
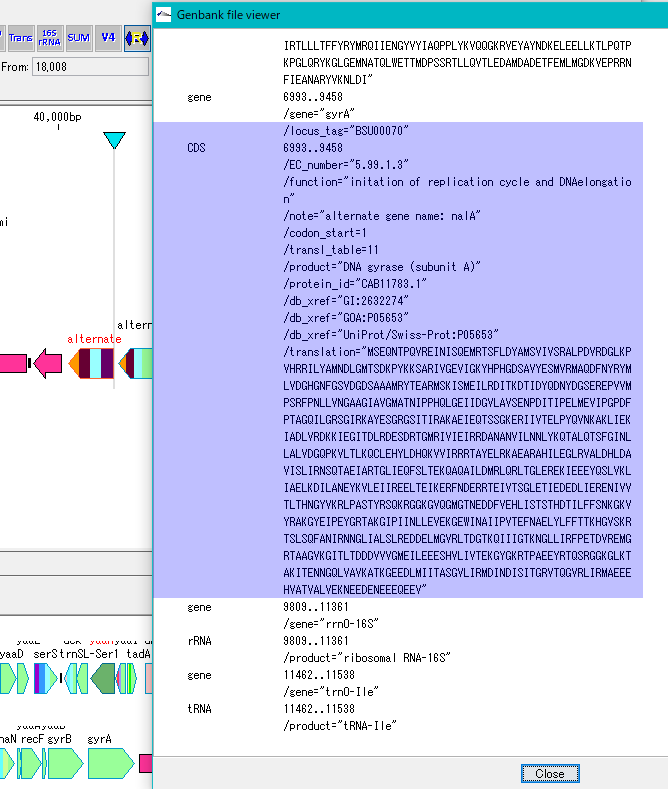
- On the other hand, if you right-click on this feature and select the Description Window from the menu and activate the Description Window, the position of this feature has been changed to the reverse complementary position.
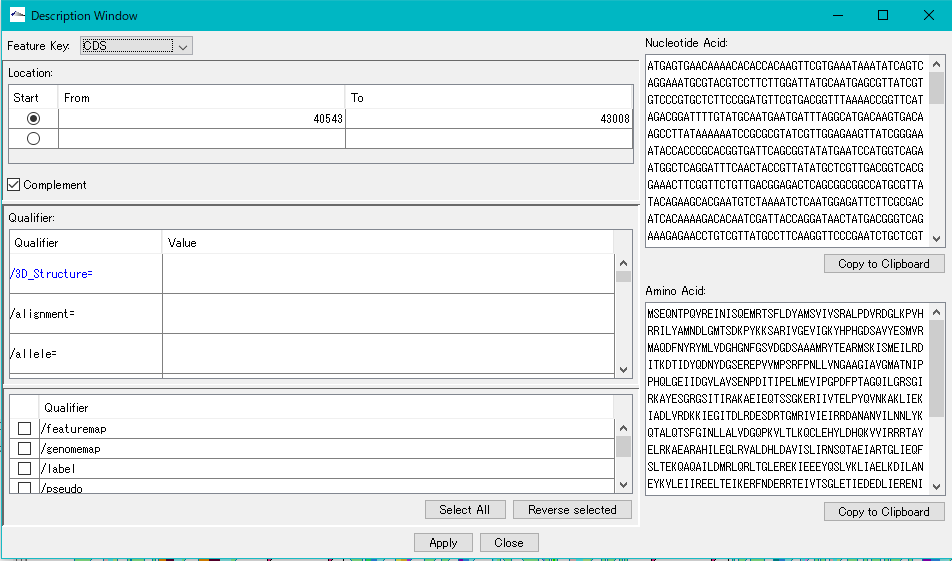
- This is because the information in the GenBank Viewer shows the contents of the file while the information on the Description Window shows the content on the memory.
- In this state, if you overwrite and save the array file, the contents of GenBank / EMBL Viewer can also be overwritten.
- Click the Reverse Complement button which is highlighted in the tool box.
- In the main feature map, the original strand is displayed.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings