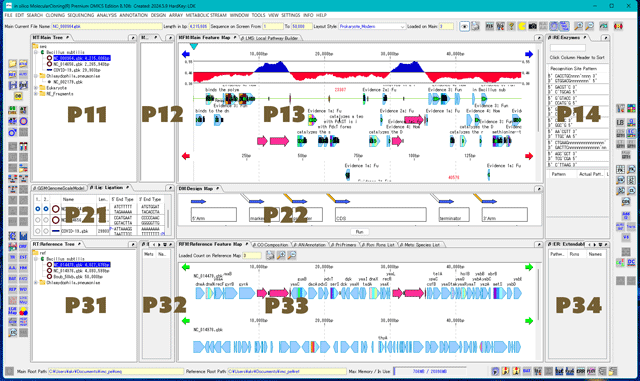
IMC N02 Overviews on Local Pathway Builder
Before this operation, execute "Set Pathway Draw Data". Execute it only once after IMC is started.
Panes used in the Local Pathway Builder
P13: 3rd column at the top: LMS: Local Pathway Builder tab pane
This is the metabolic pathway drawing area. Metabolic pathways are drawn here.
This tab pane is detachable and resizable.
P21: 1st column at the middle: GSM: GenomeScaleModel tab pane
P32: 2nd column at the bottom: EM: Extendable Metabolites tab pane
This tab pane displays a list of metabolites that can be reached in one reaction from the currently drawn metabolic pathway.
P33: 3rd column at the bottom: Rxn: Rxns List tab pane
This tab pane displays the Reaction List of the Current GenomeScaleModel.
You can sort by each column.
P33: 3rd row at the bottom: Mets: Species List tab pane
This tab pane displays the Metabolte(Species) List of the Current GenomeScaleModel.
P34: 4th row at the bottom: ER: Extendable Rxns tab pane
How to use
From the menu, click METABOLIC STREAM > Load GenomeScaleModel.
The Load SBML file selection dialog appears, so select the genome scale model (SBML format).
Note: The currently available genome scale model is the BiGG model.
When loading is complete, the information is displayed in a list in the Rxns List tab pane in the 3rd row at the bottom of P33.
Drawing a single reaction
Select the reaction you want to draw first from the P33: Rxns List tab pane and click it.
The reaction is drawn in the P13: Local Pathway Builder tab pane.
Add a reaction following a metabolite of the currently drawn reaction.
Clicking a metabolite displayed in the P32: Extendable Mets List tab pane will display the reaction mediated by that metabolite in the P34: Extendable Rxns List tab pane.
Clicking a reaction in the P34: Extendable Rxns List will add that reaction to P13: Local Pathway Builder.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings