IMC L01B Create Blast Search Databases
A database for local BLAST search can be generated from the obtained base sequence file or amino acid sequence file.
Usable base sequence formats: GenBank (.gb, .gbk, gbff), EMBL (.embl), FastN (.fna)
Usable amino acid sequence formats: GenPept (.gpff), FastA (.faa)
Operation
- From the menu click File -> Create Blast DB.
- The Blast DB List operation dialog is displayed.
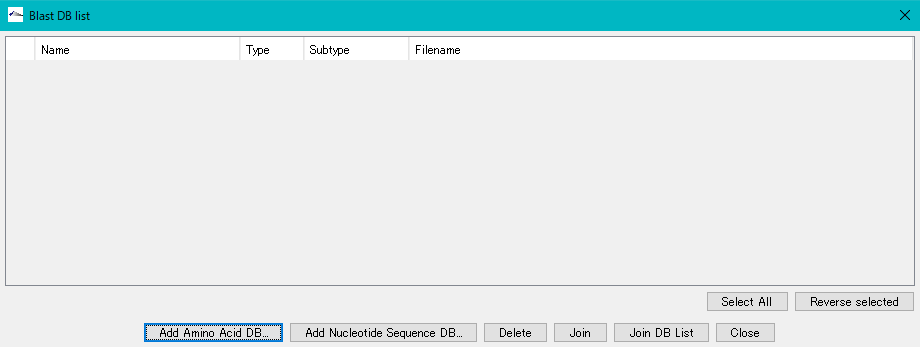
- Click Add Amino Acid DB ....
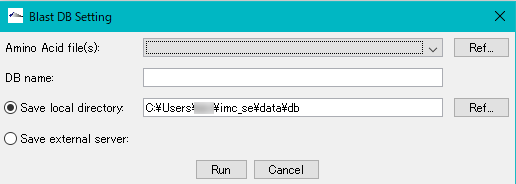
- The Blast DB Setting dialog box is displayed.
- Amino Acid File (s): Click Ref ... on the right side of the column to specify the file to create the database. It is also possible to specify multiple files simultaneously.
- The selected file will be displayed in the pull-down menu.
- DB name: In the input field, specify an arbitrary database.
- You can select either local or server storage location for the database. In order to create the database, you need to confirm in advance whether there is free space.
- The default storage location is C: \ Users \ User \ imc_xx \ data \ db on Windows.
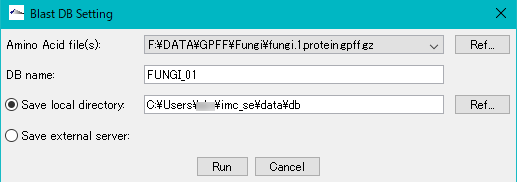
- When you click "Run", an execution confirmation message "Create Amino Acid DB?" Is displayed.
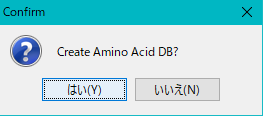
- Click [Yes (Y)] to start the database generation.
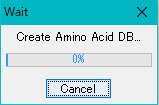
- A progress message is displayed during execution.
- If you click "Cancel" during execution, the execution will be interrupted and the database will not be created and will be terminated.
- When generation is completed, a completion message "Completed !!" is displayed.
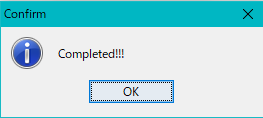
- Click "OK" to close the completion message and display the database you have created in the Blast DB List dialog.
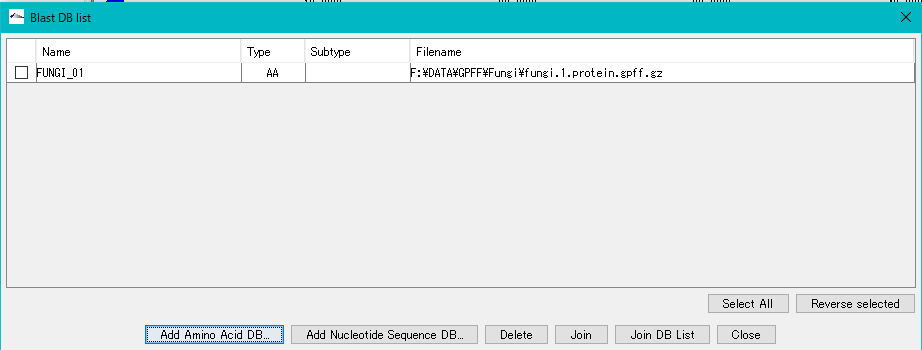
- This completes the creation of the database for Blast search.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings