IMC J04A Extract ORF Candidates from Current Genome Sequence
Extract ORF candidates from the unnotated current genome sequence.
Operation
The genome sequence from which ORF is extracted is displayed as the current sequence in the main feature map.
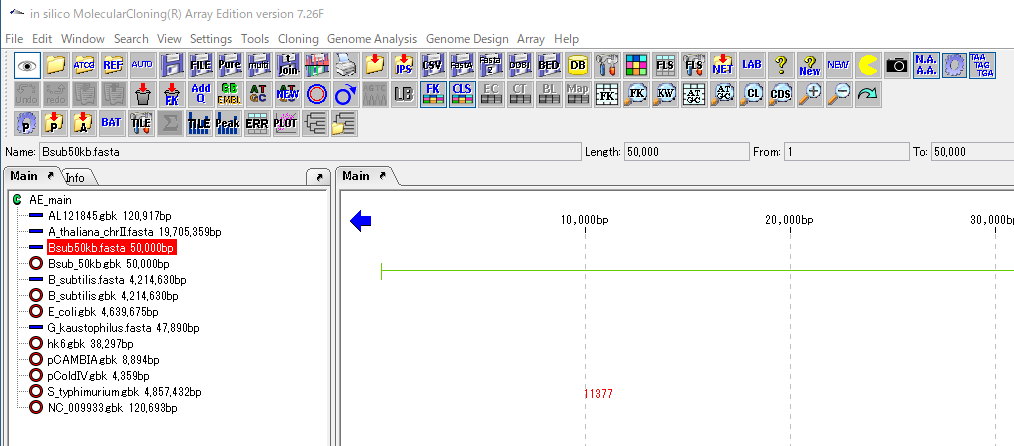
From the menu click Genome Analysis -> ORF -> ORF Extraction ....
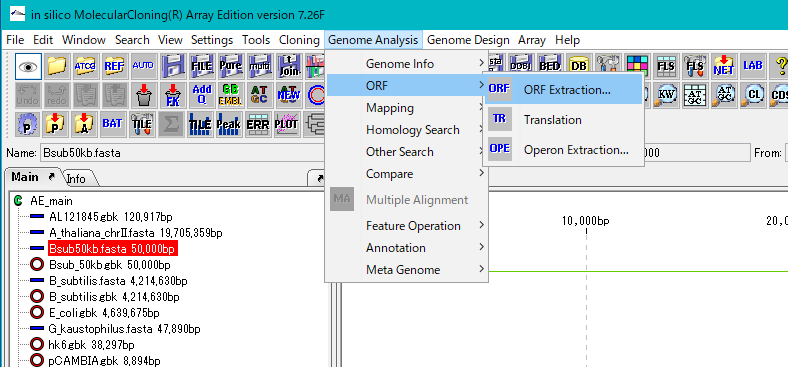
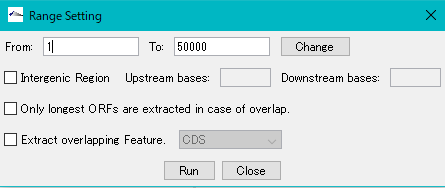
The "Range Setting" dialog will be displayed.
Check "Only longest ORFs are extracted in case of overlap".
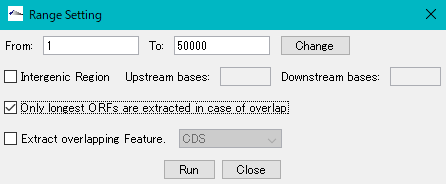
Click "Run".
A confirmation message "Start ORF Search?" is displayed.
Click "Yes".
The "ORF Site" dialog is displayed.
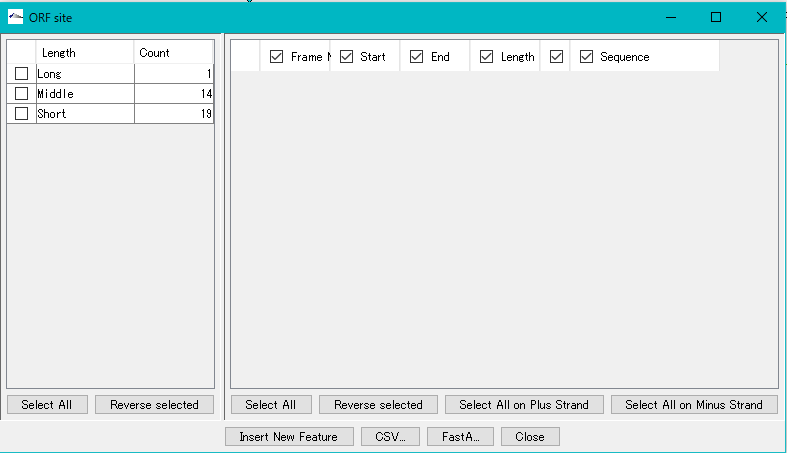
The ORF candidate extracted is classified into three types, Long, Middle and Short, depending on its base length.
You can change the range of Long, Middle, Short base length range here.
In the left pane, the number of extracted ORFs by this base length is displayed.
If you check the checkbox in the left pane, ORF candidates belonging to the base length classification checked in the right pane are displayed.
It is also possible to check only the long ORF candidates at first.
The button on the lower side of the right pane is a button for selecting a check box.
Click "Select All".
All ORF candidates in the right pane are checked.
Click "Insert New Feature".
A confirmation message "Insert New Feature?" is displayed.
Click "Yes".
The ORF candidate is registered in the current genome sequence.
A completion message is displayed.
Click "OK".
ORF candidates are registered in the main feature map and displayed.
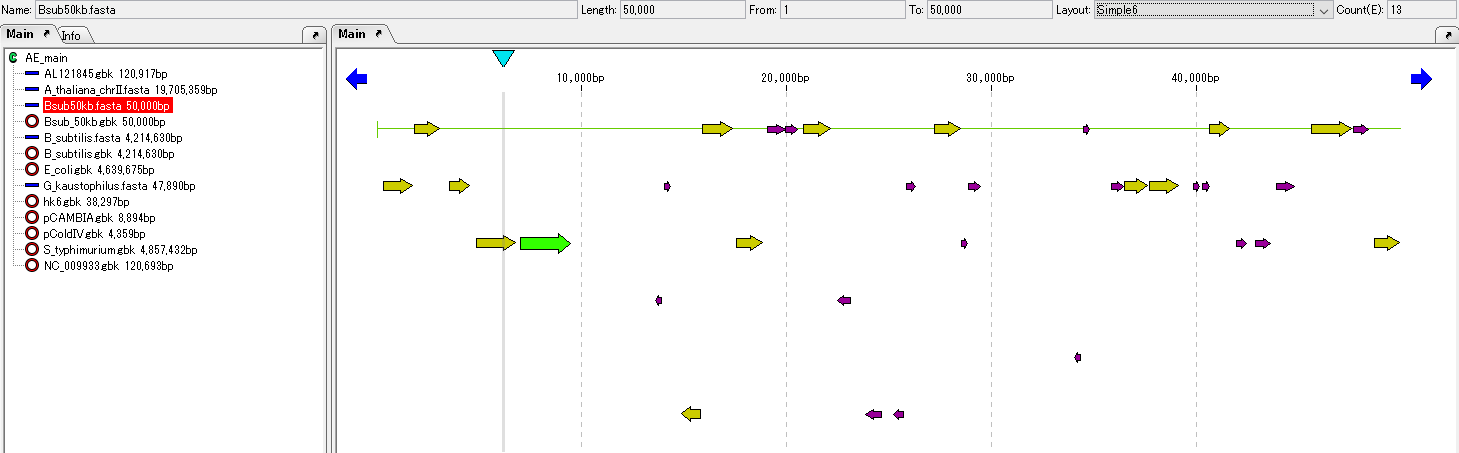
If the "ORF Site" dialog is open, clicking on one of the ORF candidate lines highlights the corresponding ORF candidate feature on the feature map and automatically scrolls the feature map horizontally (possibly not scrolling).
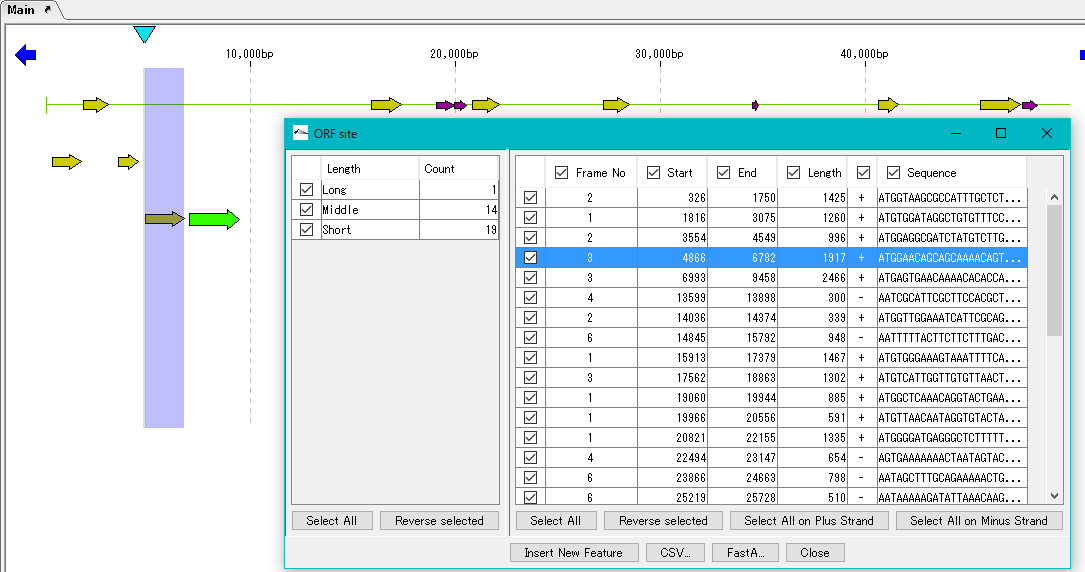
If you want to save the information of the registered ORF candidate, save using the "CSV ..." or "Fast A ..." button.
At this time, only the items checked in the checkboxes of each column are saved as files.
From the menu click File -> Save ... to overwrite the current genome sequence.
At this time, FastA file is automatically converted to GenBank or EMBL file.
If you exit without saving, the ORF candidate feature registration will be discarded.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings