GT D5A Data Format Conversion from AFG to BAM
Velvet assembler output file Convert AFG format file to BAM format file.
There are two ways.
First, if there is an AFG file loaded in the Analysis node of GT's TREE pane, you can perform conversion directly from that node.
The other is executed from GT's Tools menu.
This section explains how to perform conversion from an Analysis node.
Implementation Edition
GenomeTraveler![]()
Operation
- This is explained using training data.
- Double-click the Analysis node in the Velvet assembly.
- The afg file node is displayed.
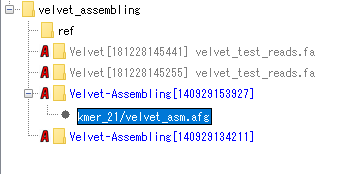
- Right-click on the afg file node on the mouse.
- The menu will be displayed.
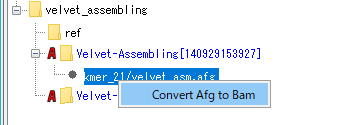
- Select "Convert Afg to Bam".
- A confirmation message will be displayed.
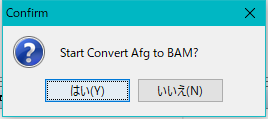
- Click "Yes (Y)".
- The conversion is executed.
- A progress message is displayed during execution.
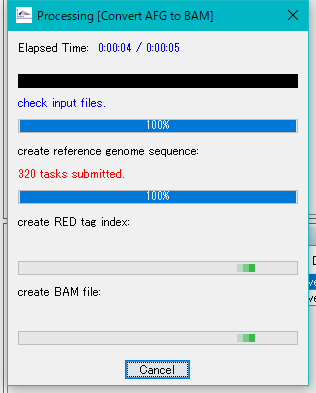
- When execution is completed, a completion message is displayed.
- Click "OK".
- The completion message closes.
- The node file has been converted to BAM.
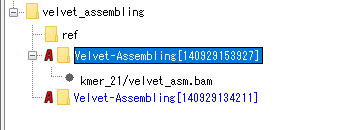
- Right-click on the BAM file node on the mouse.
- The menu will be displayed.
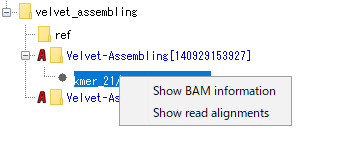
- Launch Alignment Viewer "Show Read Alignment" and display BAM information "Show BAM Information" are possible.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings