IMC O13A Operation of Mutation Search
Detects one base mutation between closely related genomes and generates a list of all mutations. In addition, from the list you can display the variation of the reference feature map and the base position of the corresponding main feature map.
Preparation
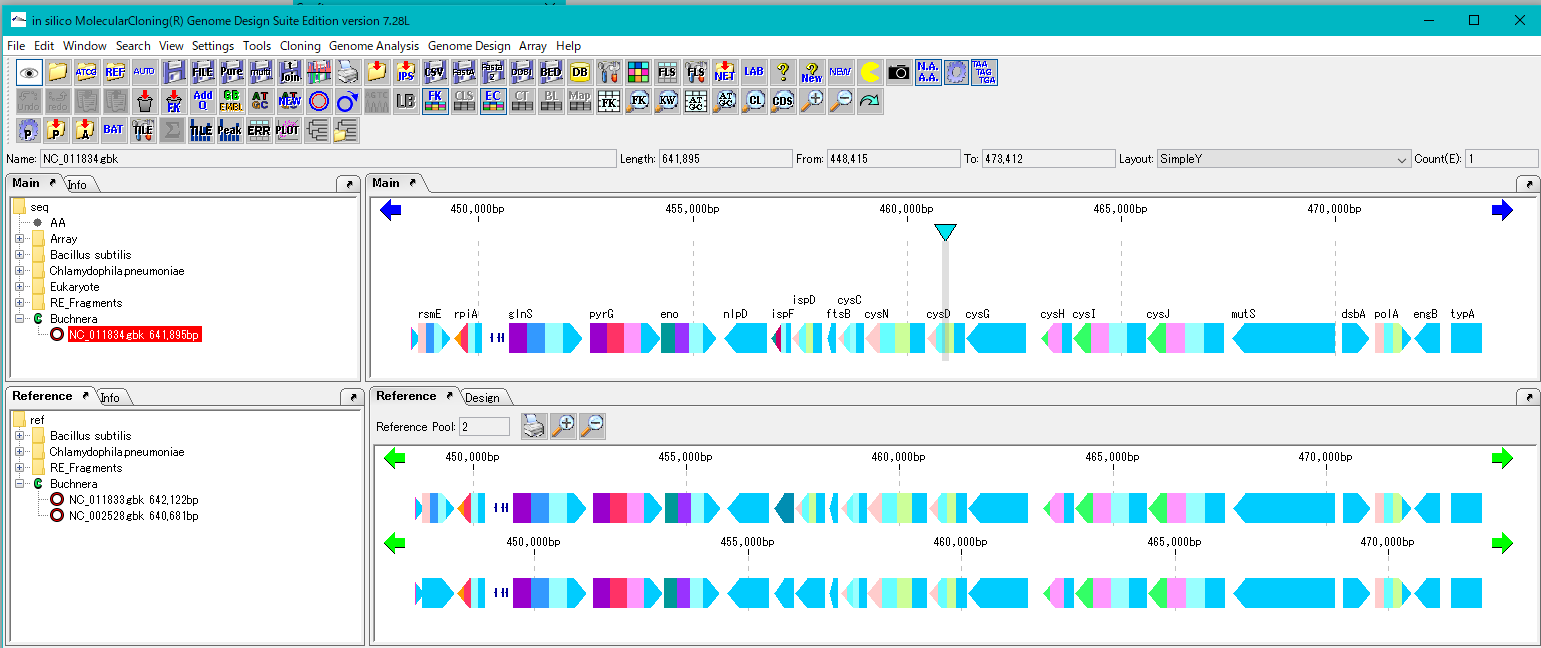
Load the base stock genome into the current main feature map.
Load multiple closely related genomes to compare with the current reference feature map.
Operation
Select Genome Analysis -> Compare -> Mutation Search from the menu.
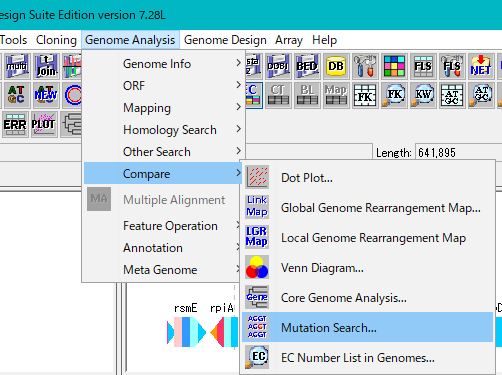
The Mutation Search Setting Dialog Box will be displayed.
All genomes loaded in the current reference feature map are listed.
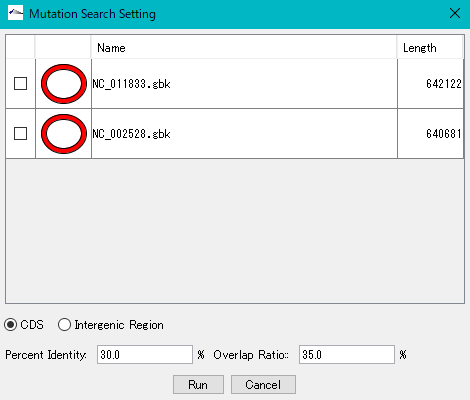
Check the genome (multiple designation possible) to extract the mutation.
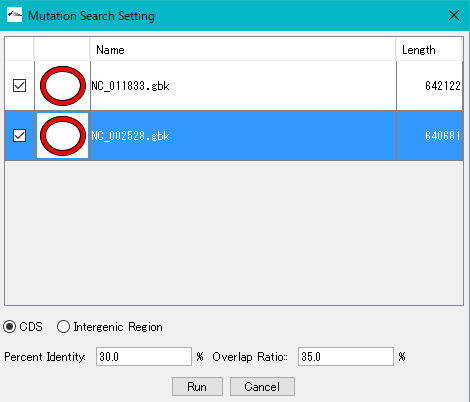
Click "Run".
A confirmation message "Start Mutation Search?" Is displayed.
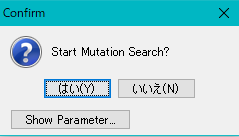
To change the execution parameters, click the Show Parameter button.
Click "Yes (Y)".
Execution is started and a progress message is displayed during execution.
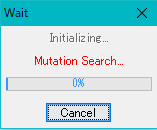
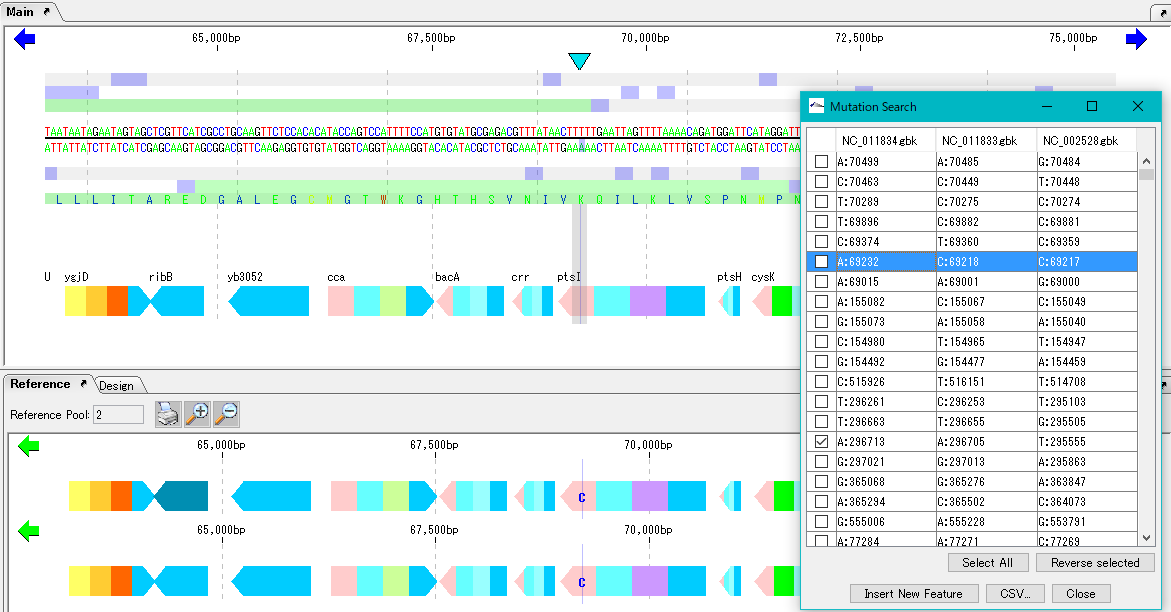
When execution is completed, the list of detected mutations is displayed.
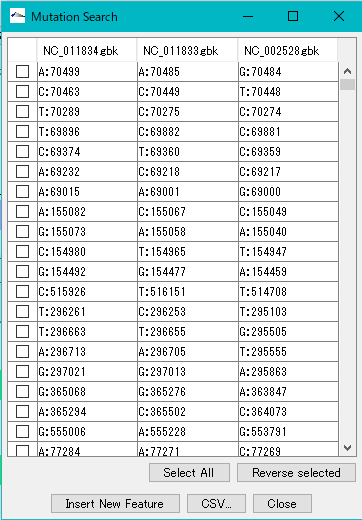
In this list, the mutation points detected in each row are displayed. The mutation point shows the base position by genome and its base call.
When you click on one row (mutation point), the main feature map and the reference feature map are automatically shifted, and the mutation part is displayed as the center of the map.
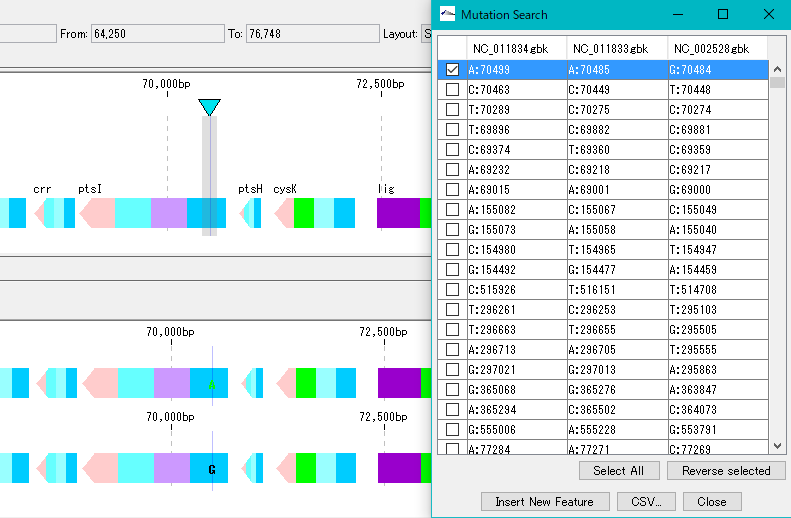
Mutable bases are highlighted on the sequence lane of the main feature map.
On the other hand, in the reference feature map, the variant base is displayed at the mutation site.

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings