GT 11D Practice Fragment Mapping using Training Data
Practice the fragment mapping operation using training data.
Operation
Start GT.
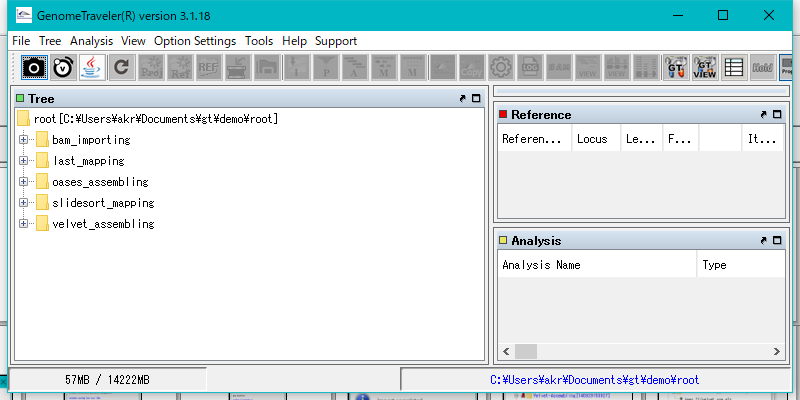
Right click mouse over "last_mapping" node.
The menu will be displayed.
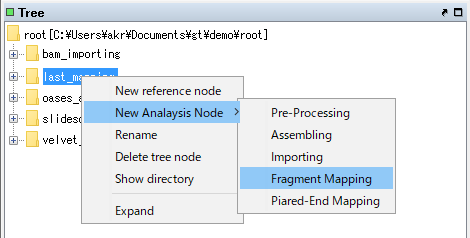
Select "New Analysis Node -> Fragment Mapping".
The Fragment Mapping dialog will be displayed.
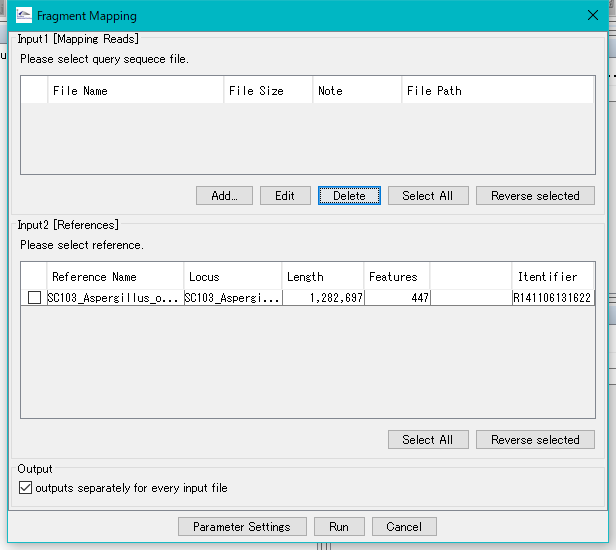
Click Add ... and select a file naeded *demo.fasta from the demo data (training data).
Check all check boxes.
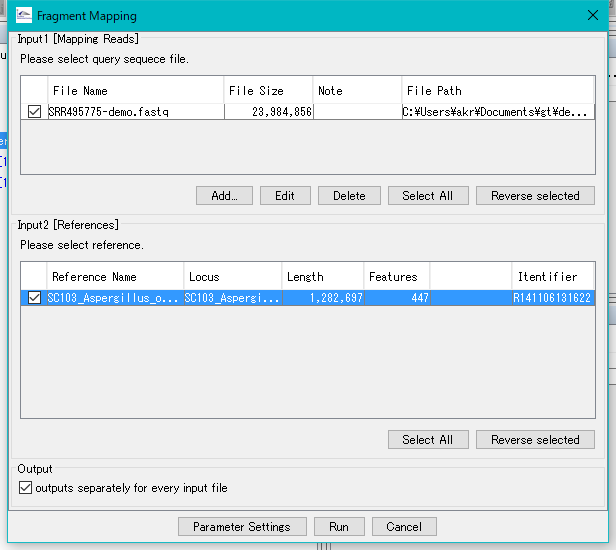
Click Run.
An execution confirmation message is displayed.
Click "Yes (Y)".
Execution is started and a progress message is displayed during execution.
When the mapping process is complete, a completion message is displayed.
Click "OK".
The node of this mapping result is created at the top of Analysis node of last_mapping node.
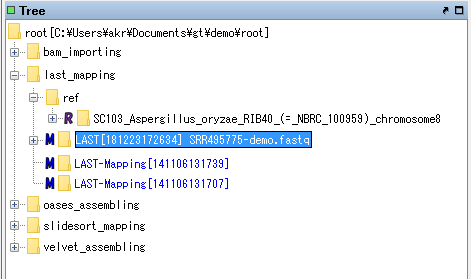
Right-click on the node and select Expand from the displayed menu, or double-click the node.
The bam file node is displayed below the node.
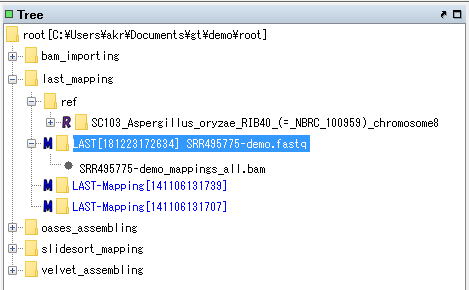
Right-click on the bam file node on the mouse.
The menu will be displayed.
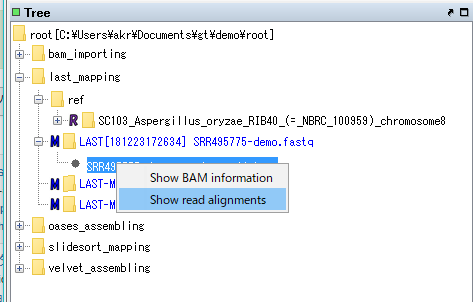
Select "Show Read Alignment".
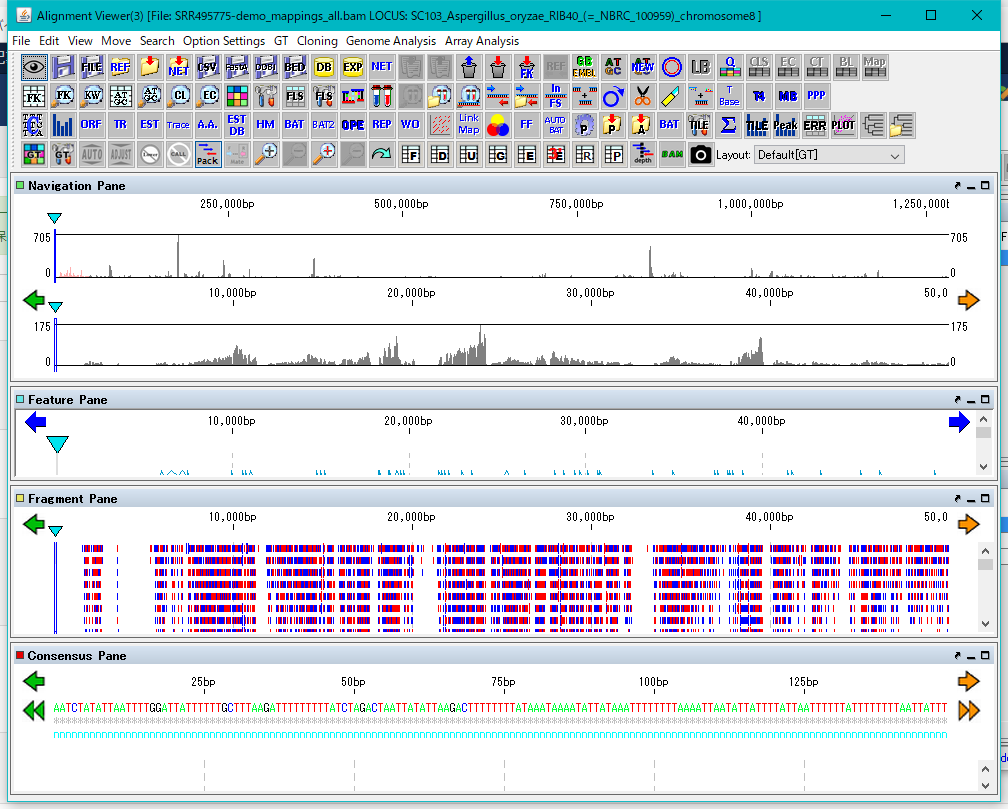
The Alignment Viewer for viewing and analyzing the mapping result is displayed.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings