IMC i01C Restriction Enzyme Digestion of DNA Sequence and Load Fragment
Digestion of the current DNA sequence with restriction enzyme and loading the restriction fragments.
Operation
- Load the DNA sequence file.
- For example, load "Bsub_50 kb.gbk".
- The feature map is displayed.
- Select "Cloning -> Restriction Enzyme -> RE Registration & Editing" from the menu.
- The "Enzyme Selection" dialog is displayed.
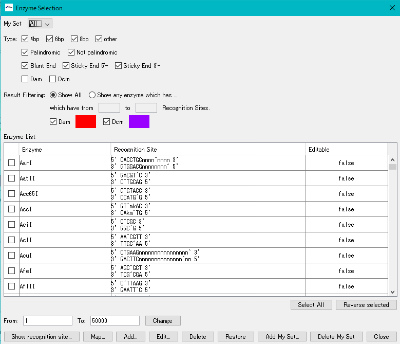
- Click the "Select All" button.
- All restriction enzymes are checked.
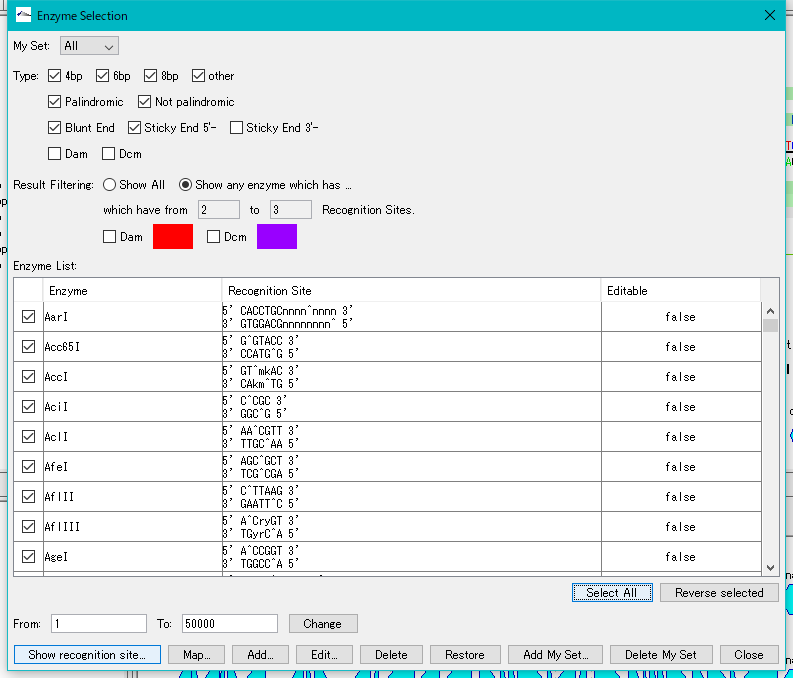
- Click the "Show Recognition Site" button.
- The Recognition Site window is displayed.
- The restriction enzyme name and the number of recognition sites detected are listed in the left pane.
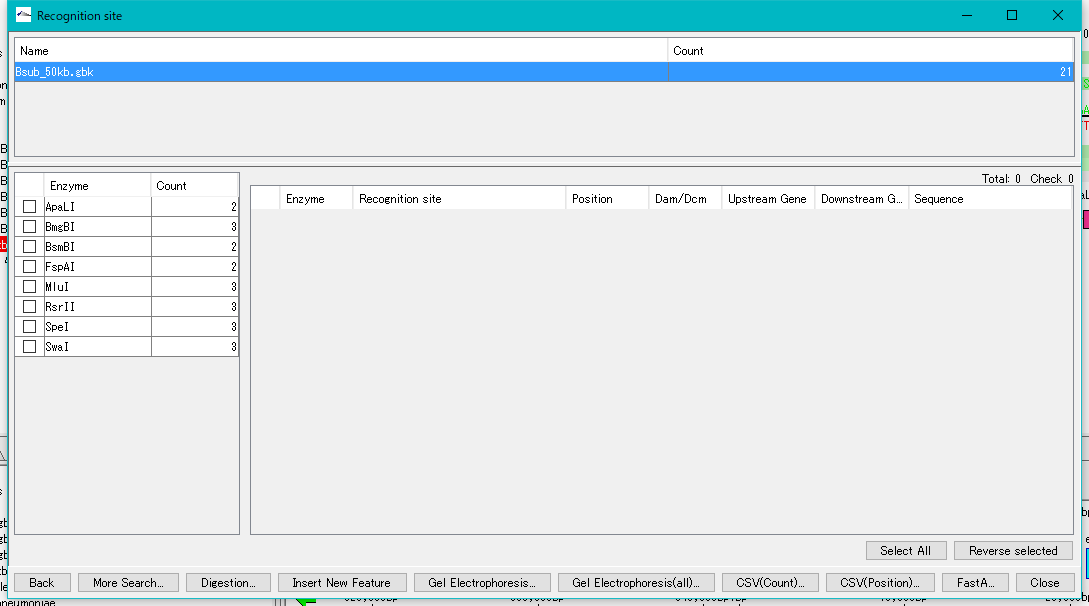
- Check with restriction enzyme (multiple selection is possible).
- The recognition site of the selected restriction enzyme is listed in the right pane.
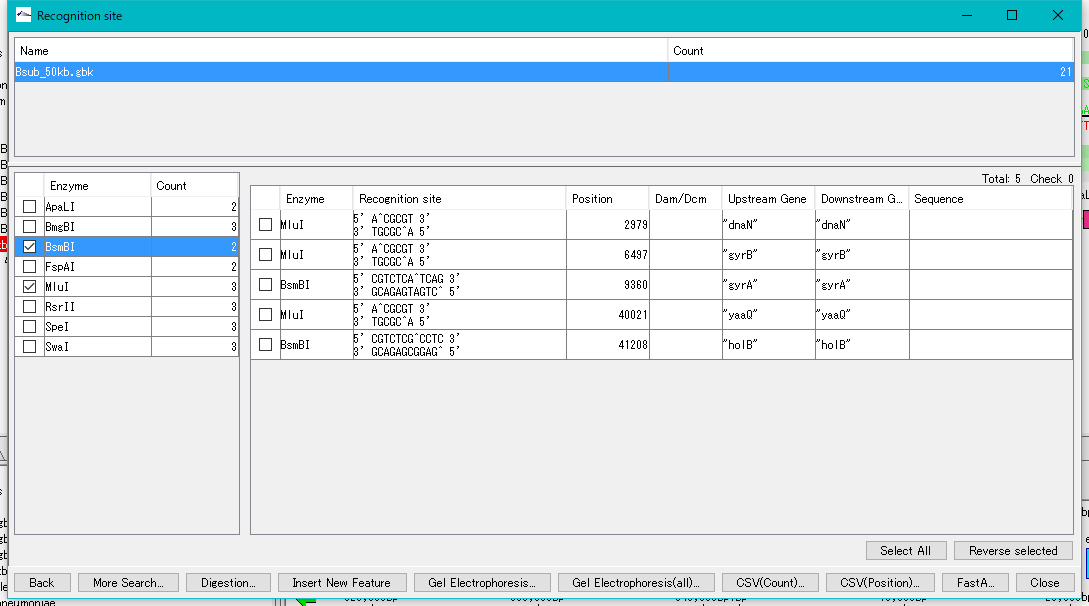
- Click "Select All".
- All check boxes in the right pane are checked.
- Click "Digestion ...".
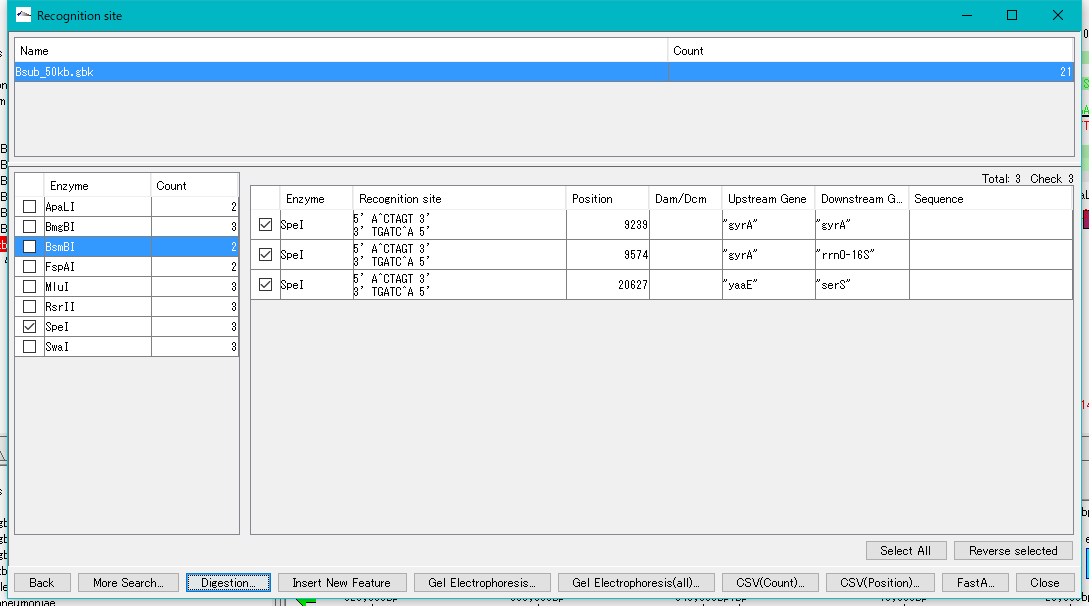
- A "Digestion?" Confirmation message will be displayed.
- Click "Yes (Y)".
- The "Digestion List" dialog will be displayed.
- In this list, file name automatically appended to digestion cut piece, base length, position on original DNA sequence, name of restriction enzyme site at both ends, actual recognition site sequence, DNA sequence and the like are displayed I will.
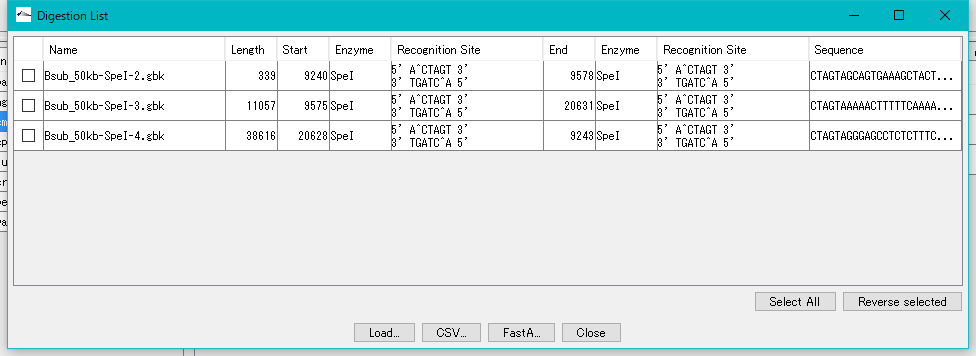
- Click the "Select All" button.
- All fragments are checked.
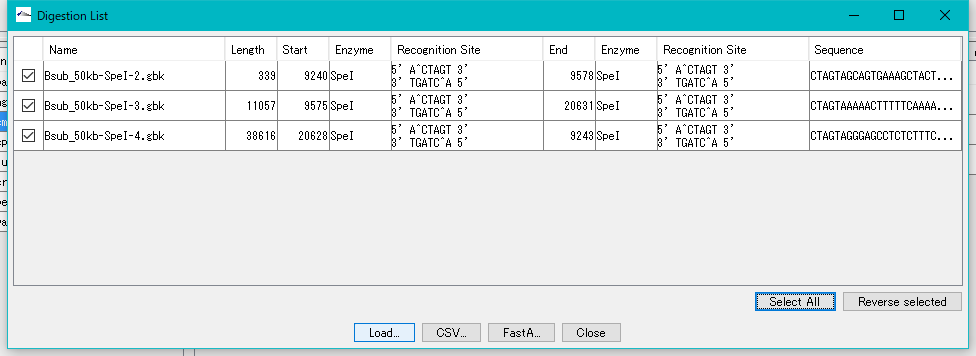
- Click "Load ...".
- A completion message is displayed.
- Restriction fragments are loaded into the current main directory.
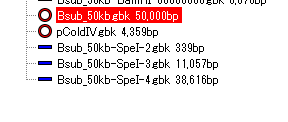
- Click on one of the loaded nodes.
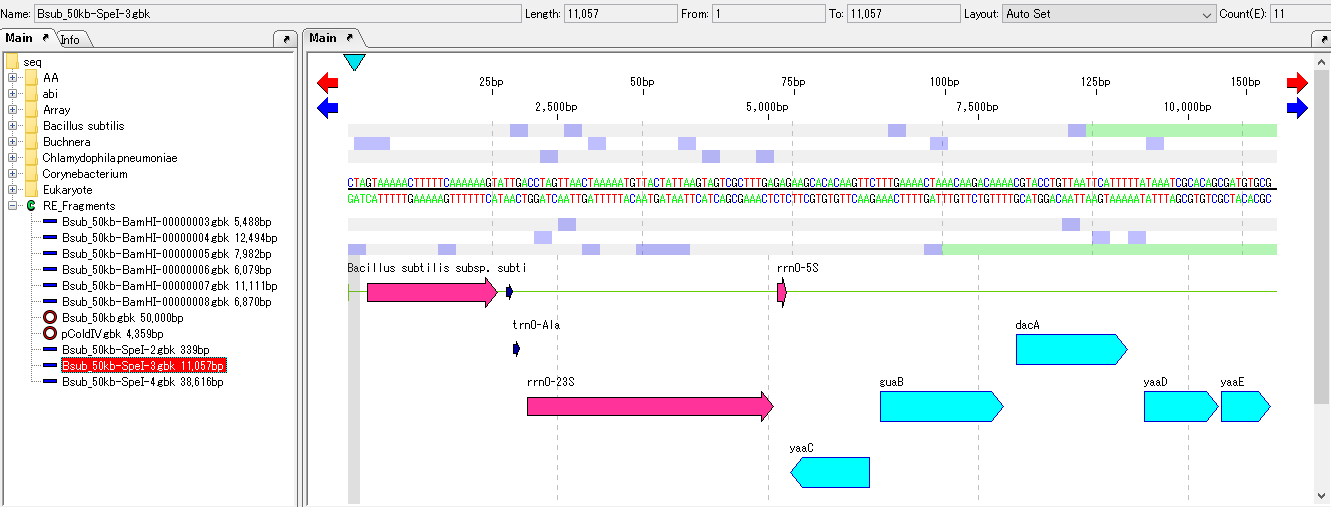
- A feature map of restriction enzyme digested fragments is displayed on the main feature map.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings