IMC W708 Blast Color Setting Tab Pane Description
In the Blast Color tab pane, as a result of executing batch homology search and automatic annotation, we set up to color classify each feature according to the items automatically posted to the feature.
There are two types of color classification, one to solidly paint each feature figure with one color, and the other to change the fill color depending on the degree of match for each base sequence or amino acid sequence of the feature.
In the latter case, the paint color will be thinned out on a large scale.
To select from two types, use the View Type radio button.
View Type Radio Button: When Area is turned on, paint the entire feature with one color.
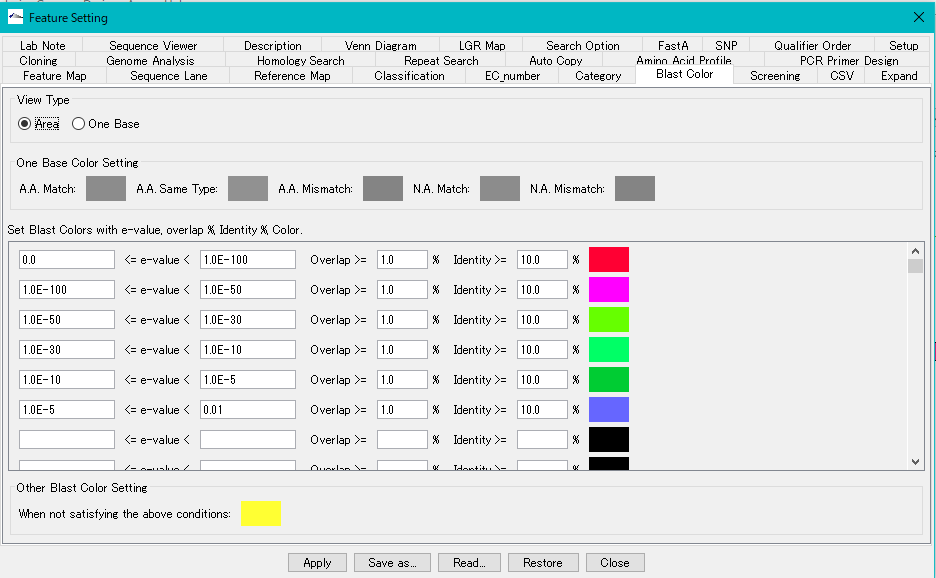
Set Blast Color with e-value, overlap,% identity and Color Section:
In this section, you set the fill color according to the condition of the homology score. Conditions can be set up to 100 conditions, conditions are examined from the top of the list. When the condition of the line is satisfied, the fill color is applied, and the condition lower than it is not judged. With respect to the line for which no judgment condition is entered, no judgment is made and the process moves to the lower condition judgment as it is. If all the conditions are not satisfied, the setting in the "Other Blast Color Setting" section is applied.
- E-value range text field: For E-Value, set upper and lower range of condition. Enter zero or positive real number. Enter a value less than the value on the right side of the value on the left side. Exponential notation is also possible.
- Overlap text field: Specify the ratio of the alignment length of the query array and the subject array to the total length. Enter a positive real number.
- % Identity text field: Specify the degree of matching of bases or residues in the alignment region of the query sequence and the subject sequence as a percentage. Enter a positive real number.
- Other Blast Color Setting section: Here we set the fill color when not satisfying all of the condition list.
View Type radio button: When One Base is turned on, change the fill color with the degree of matching of all bases or residue units in the feature range.
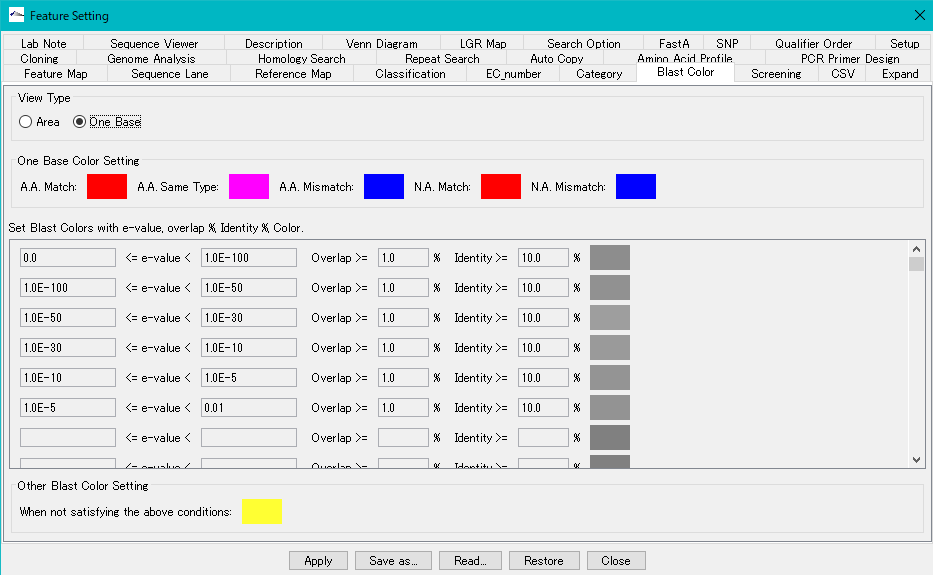
One Base Color Setting section:
As a result of the homology search, you can change the filling car by whether the query sequence and the subject sequence match for each base or residue.
- A.A. Match color box: This fill color will be applied if the query and subject amino acids match.
- A.A. Same Type color box: This fill color is applied when query and subject amino acids are of the same type.
- A. A Mismatch color box: This fill color is applied if the amino acids of the query and the subject do not match.
- N.A. Match color box: This fill color is applied when the query and subject bases match.
- N.A. Mismatch color box: This filling color is applied if the bases of the query and the subject do not match.
For Feature Setting common operation buttons, please click here.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings