IMC W212B Blast Search Result Dialog Explained
The items on the Blast search result dialog and the operation method are explained.
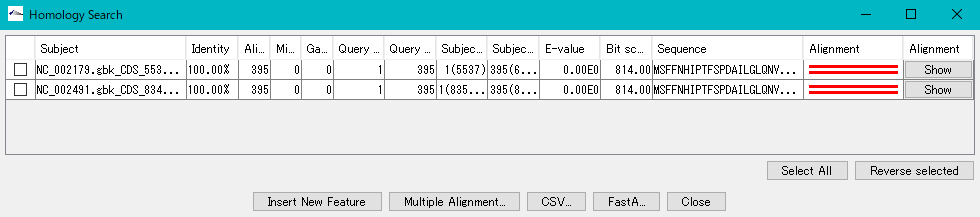
A list of hit Subjects is displayed one line at a time.
Display items
- Check box for selection
- Subject: Name of the array on the hit database (array name + feature key + array position)
- Identigy: Percent identity of the query sequence and the hit sequence on the database (Percent Identity)
- Alignment Length: Alignment base length or amino acid residue length
- Mismatches: Number of unmatched bases or amino acid residues
- Gap openings: Number of gaps
- Query start: Alignment start position on the query array
- Query end: Alignment end position in query sequence
- Subject start: Alignment start position (absolute position on genome) in hit arrangement
- Subject end: Alignment end position (absolute position on genome) in hit arrangement
- E-value: Expected value of appearance
- Bit score: The homology score determined by the value of the matrix of the permutation matrix used for the search
- Sequence: Query sequence used
- Alignment: A graphical representation of the alignment status. It is indicated by a line segment representing the query sequence and a line segment representing the hit arrangement in parallel above and below. Regions displayed in red indicate areas with homology, and areas displayed in black indicate areas with no homology.
- Alignment: A button to the alignment dialog, when clicked, an alignment dialog is displayed.操作方法
Linking search result list and feature map
- Clicking on each line in the hit list displays the position of the hit array.
- If you use the reference feature map as the search database, the reference feature map shifts so that the reference feature map moves and the hit array appears in the middle.
- When using the main feature map as the search database, the main feature map is automatically shifted so that the hit array on the main feature map is displayed in the center.
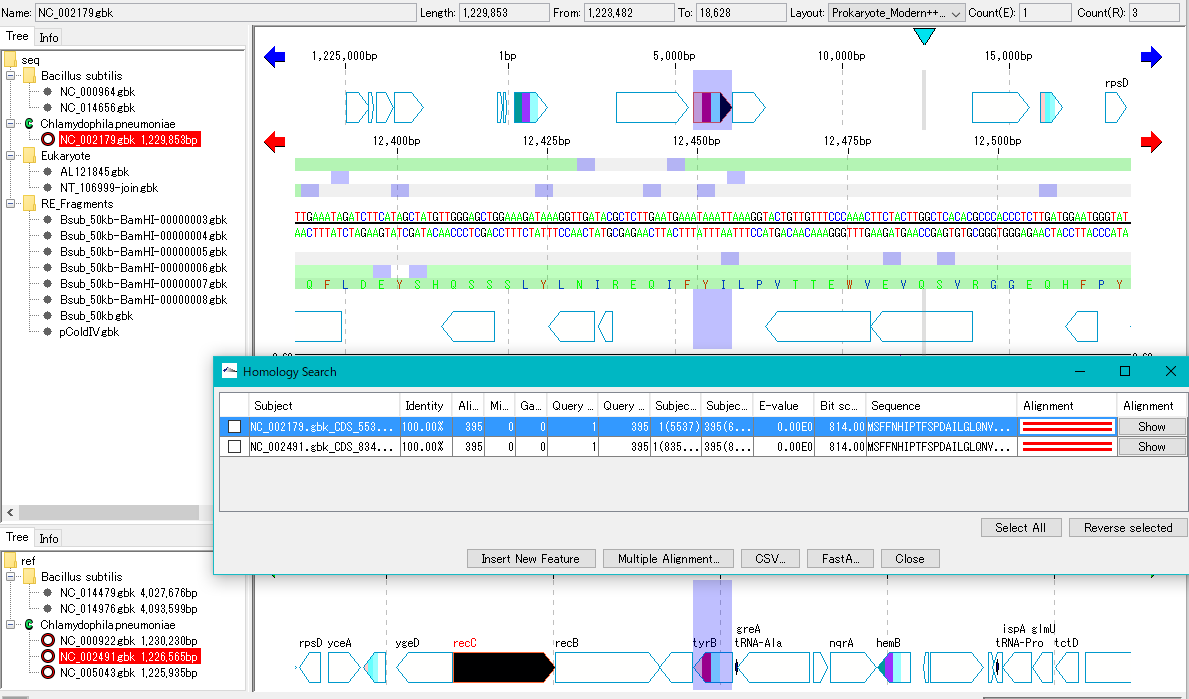
Display Alignment window
- When you click Aligment on each line of the hit list, the Alignment window is displayed.
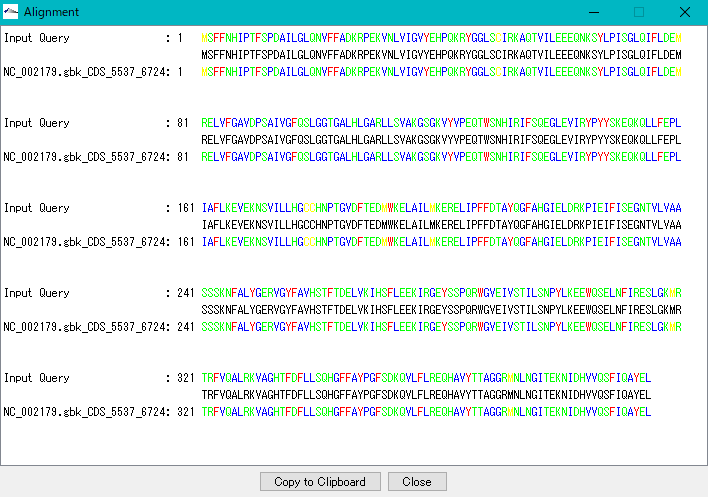
- In the alignment window, a pairwise alignment of the query sequence and the hit array is displayed.
- In the Alignement dialog, click Copy to Clipboard to copy the alignment to the clickboard.
- Click Close to close the Alignement dialog.
- Register homologous regions as new features on the current genome sequence
- Check the hit entry you want to register as a feature (multiple designation possible).
- Click "Insert New Feature".
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings