IMC W724 LGR Map Tab Pane Description
In this tab pane you can change the settings for the local genome rearrangement map.
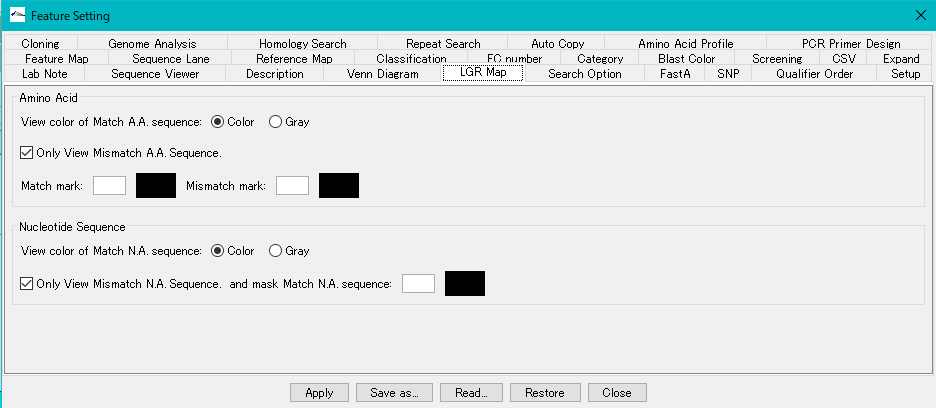
Amino Acid Section
View Color of Match A.A. Sequence Color Toggle Radio Button: When on, displays the amino acid residue characters displayed on the sequence lane in color.
View Color of Match A.A. Sequence Gray Toggle Radio Button: When on, displays the amino acid residue characters displayed on the sequence lane in grayscale.
Only View Mismatch A. A. Sequence check box: When checked, displays the amino acid residue letters only if the amino acid residues of the two genomes being compared match. When unchecked, all amino acid residues are displayed.
Match Mark input field: Specify the character to be used in the event that the amino acid residue matches.
Match Mark color box: Specify the character color to be used in the event that amino acid residues match.
Match Mark input field: Specify the character to be used in the event that the amino acid residue does not match.
Match Mark color box: Specify the character color to use when the amino acid residues do not match.
Nucleotide section
View Color of Match Nucleotide Color Toggle Radio Button: When on, the base letters displayed on the sequence lane are displayed in color.
View Color of Match Nucleitide Gray Toggle Radio Button: When on, displays the base letters displayed on the sequence lane in grayscale.
Only view Mismatch Nucleotide and Mask Match Nucleotide check box: When checked, the base letters are displayed only when the bases of the two genomes to be compared match. When unchecked, all base letters are displayed.
Input field: If the bases do not match, specify the character to use during that time.
Color Box: Specify the character color to be used if the bases do not match.
For Feature Setting common operation buttons, please click here.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings