IMC S411 Execute OGAB Fragmentation Designer
To rebuild the designed gene cluster, fragment the cluster optimally.
After each fragment is chemically synthesized, each fragment is reconstituted by OGAB method to prepare a long chain DNA cluster.
Implementation Edition
This function is implemented in the following edition.
IMCDS![]()
Operation
- Select Genome Design -> OGAB Fragment Design from the menu.
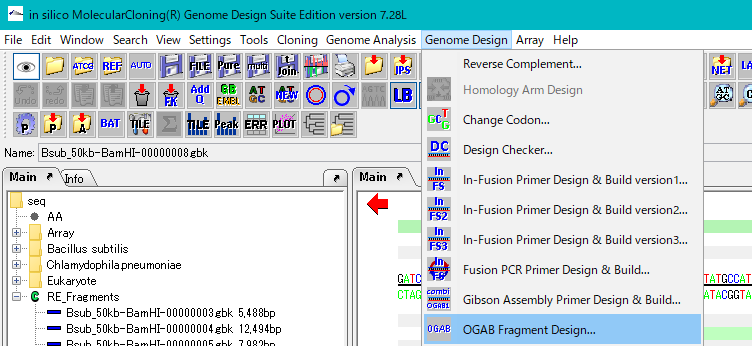
- The "OGAB Method" execution dialog is displayed.
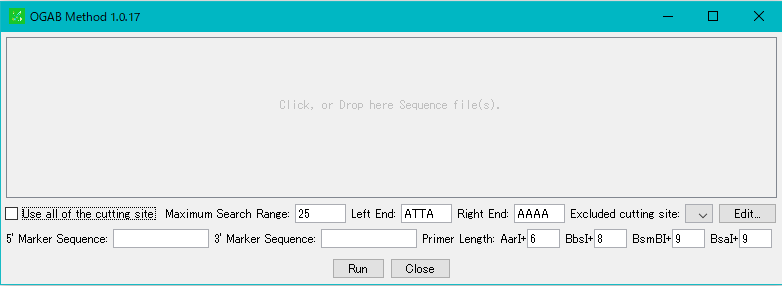
- Drop the designed cluster sequence file in the drop area or click on the drop area to display the file selection dialog and select the cluster sequence file.
- GenBank / EMBL format, FastA format cluster sequence can be specified.
- Multiple sequences can be specified.
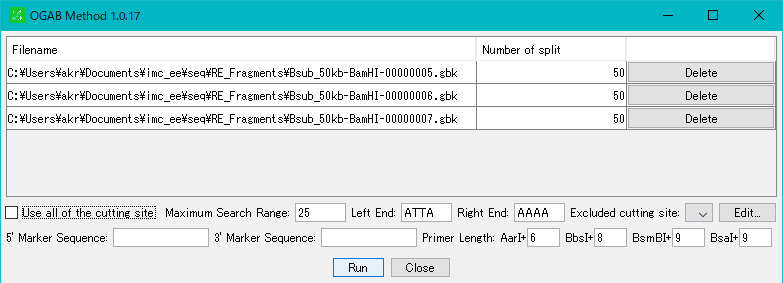
- Click "Run".
- The fragment design is executed and a progress message is displayed during execution.
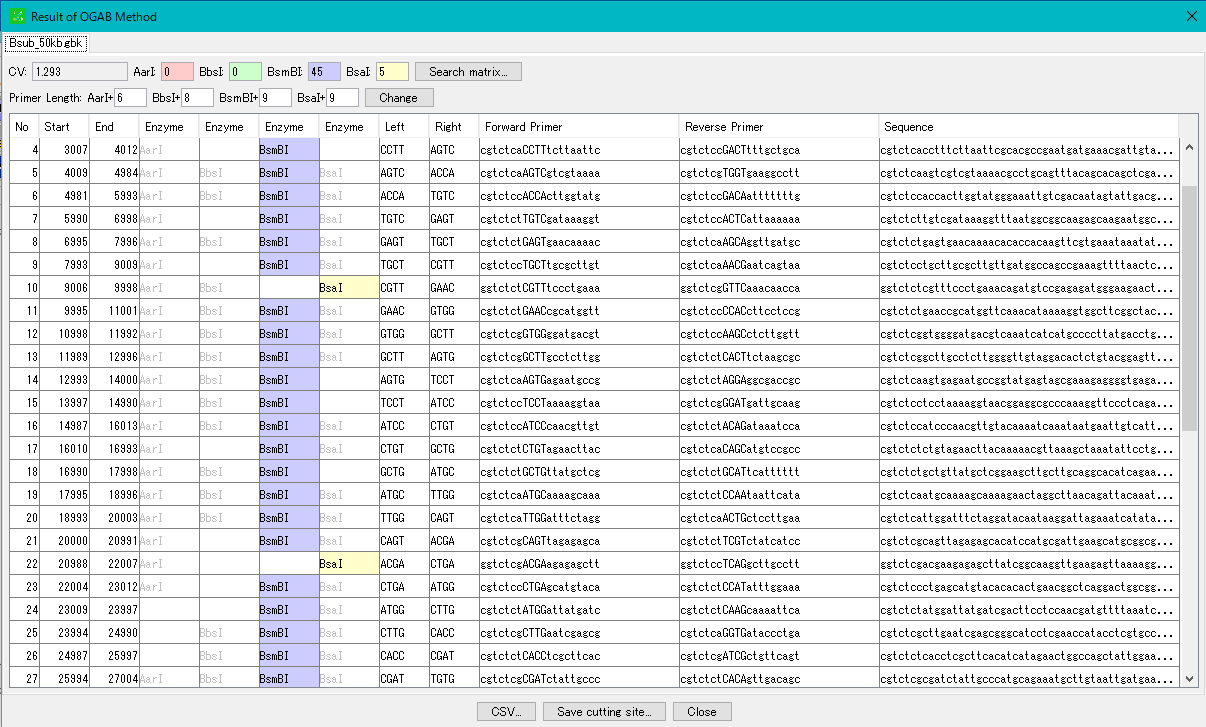
- When execution is completed, the "OGAB Fragmentation Result" dialog will be displayed.
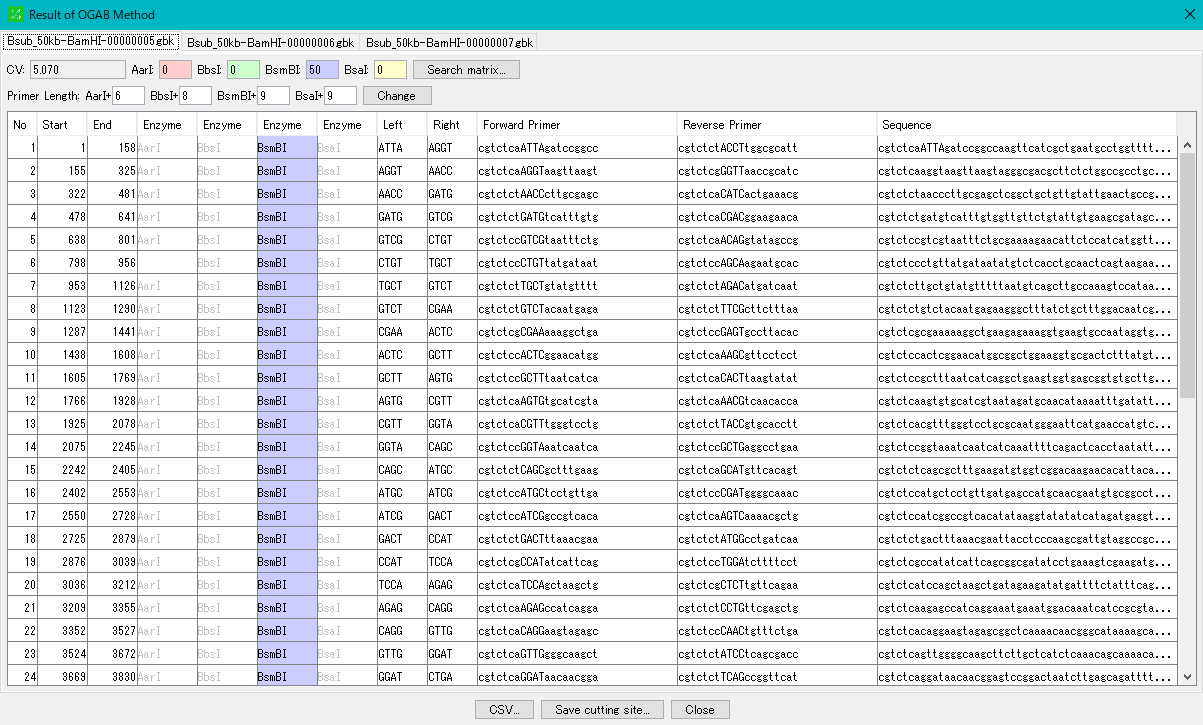
- In the result dialog, a tab pane is generated for each cluster sequence to be executed, and you can display each result pane by clicking the tab.
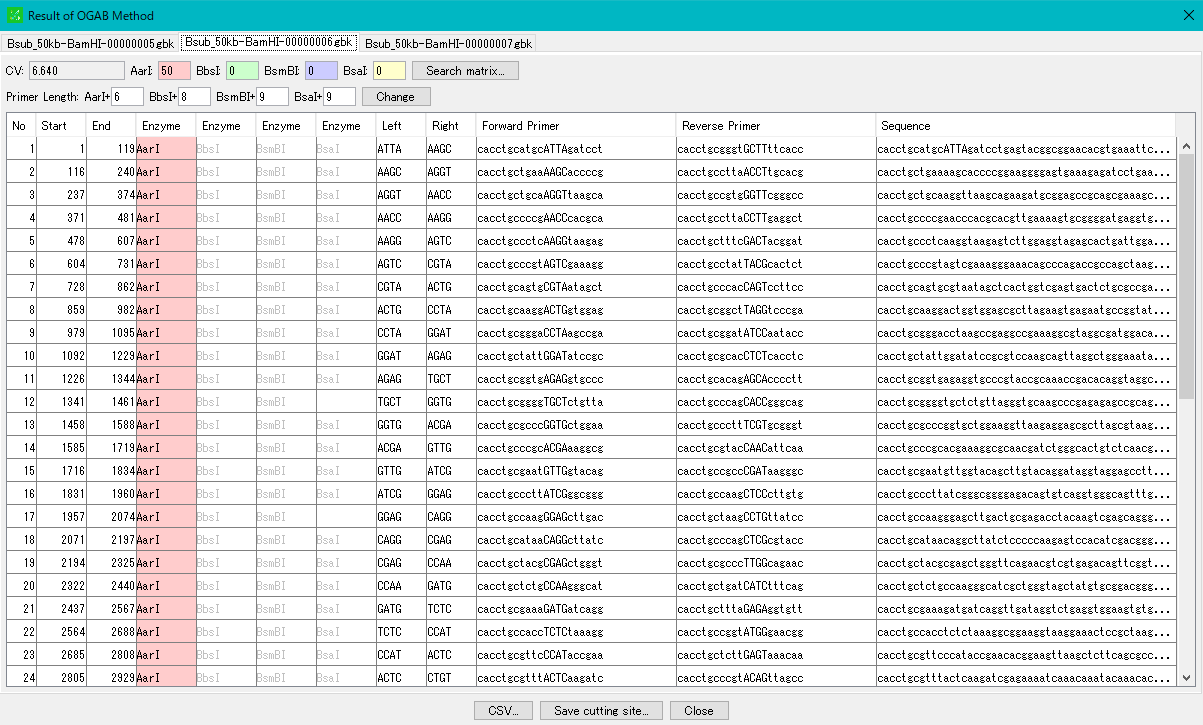
- For details on how to operate the "OGAB Fragmentation Result" dialog, please click here.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings