IMC W721 Sequence Viewer Tab Pane Description
In the Sequence Viewer tab pane, you can change display settings of Sequence Viewer.
The Sequence Viewer complements the sequence lane of the main feature map.
Traditional sequence analysis software targets users who are using those functions because there are many formats that display the nucleotide sequence with a new line.
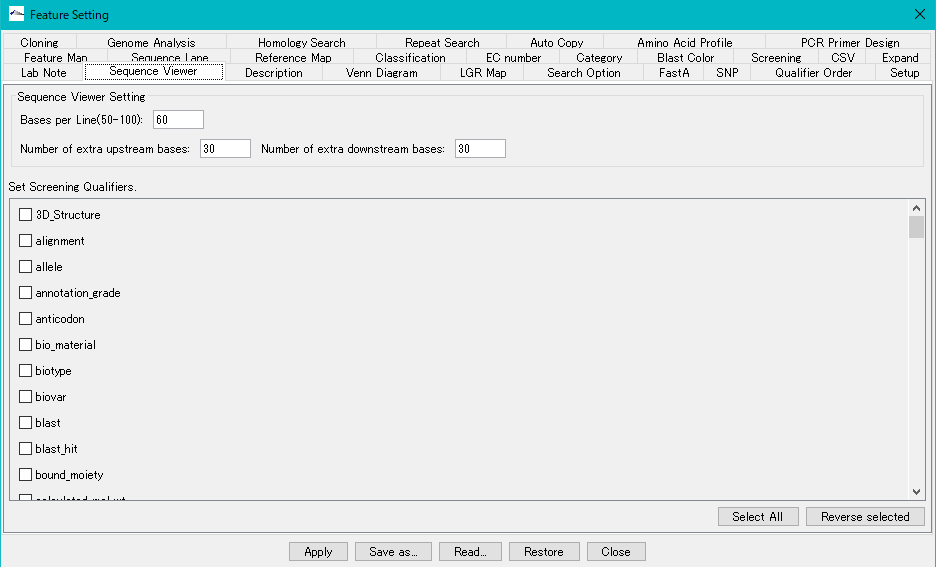
Sequence Viewer Setting section
Base per Line (50-100) Text field: Enter an integer between 50 and 100. A new line is specified with the number of bases specified here.
Number of Extra Upstream Bases Input field: Enter a positive integer. This number specifies the number of bases to cut out and output an extra stream upstream from the specified area when cutting out the sequence.
Number of Extra Downstream Bases Input field: Enter a positive integer. This number specifies the number of bases to extract and output an extra stream downstream from the specified area when cutting out the sequence.
Set Screening Qualifiers list
Remove the checked Qualifier from the output file.
Select All button: Click to check all Qualifiers.
Reverse Selected button: When clicked, unchecks the Qualifier being checked and checks unqualified Qualifiers.
For Feature Setting common operation buttons, please click here.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings