IMC 021 Make Reverse Complement of Genome Sequence in a Moment
Funtion
The current sequence can be easily switched to reverse complement display. For the base sequence, Complement of the last base becomes the first base, and the display position of each feature is also reversed all the way.
When saved as a file in this state, the base sequence is recorded in reverse complementary strand, all the positions of all the features are converted to the position from the end of the sequence and rewritten to belong to the opposite strand.
Operation
The current sequence is the target.
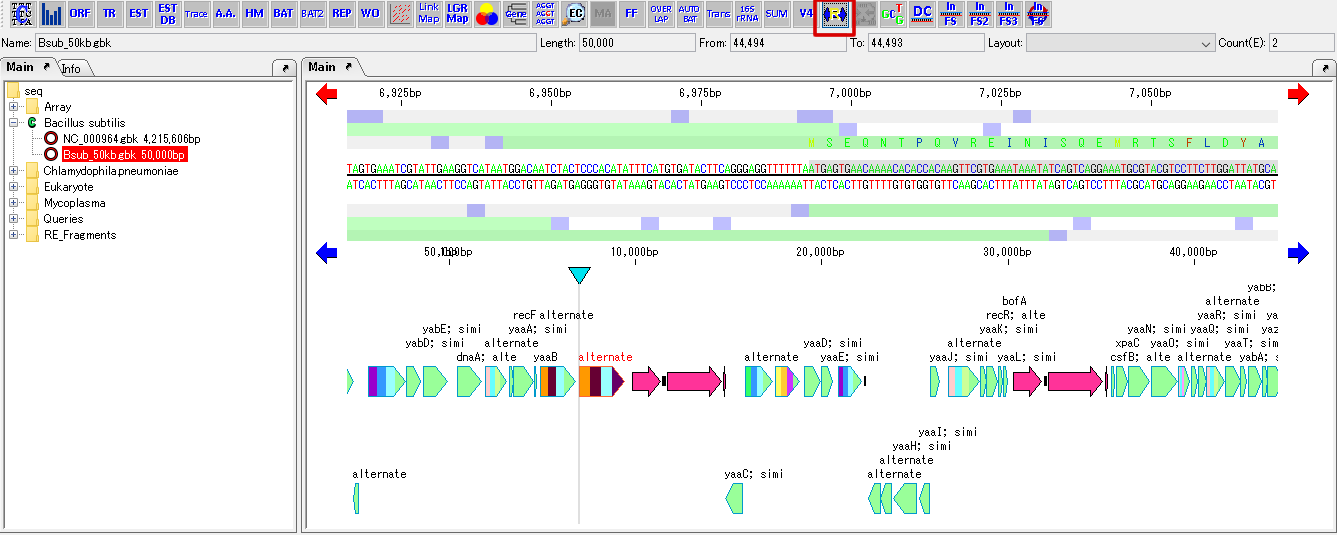
- From the menu, select Genome Design -> Reverse Complement.
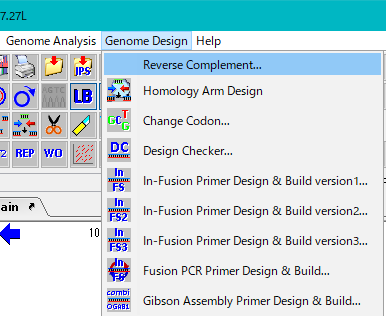
- If there is a check in the submenu, this indicates that the reverse complementary strand is already displayed from the original sequence.
- The whole is reverse complementary strand display.
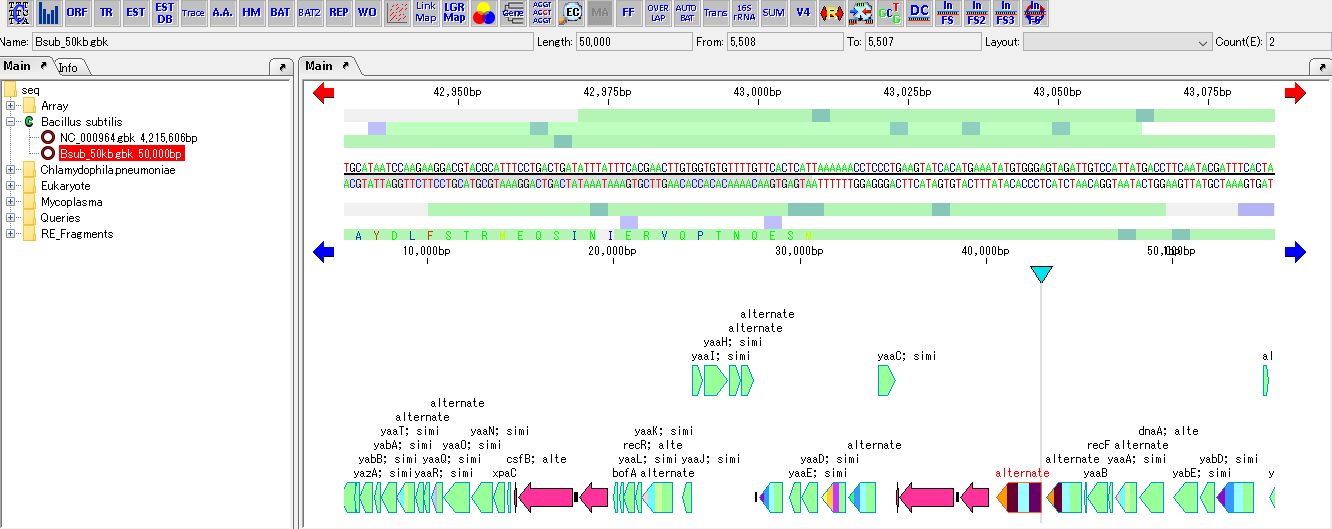
- When saved in this state or saved under a different name, the saved file is written in reverse complementary strand.
- If you do the same operation again, it returns to the original.

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings