IMC W33A Amino Acid Profile's Motif List Setting Dialog
At the top of the amino acid profile viewer, the amino acid sequence is displayed but you can display in highlight color the amino acid sequence motifs detected in it.
Amino Acid Profile (s) The Motif List setting dialog registers and manages the motifs displayed there.
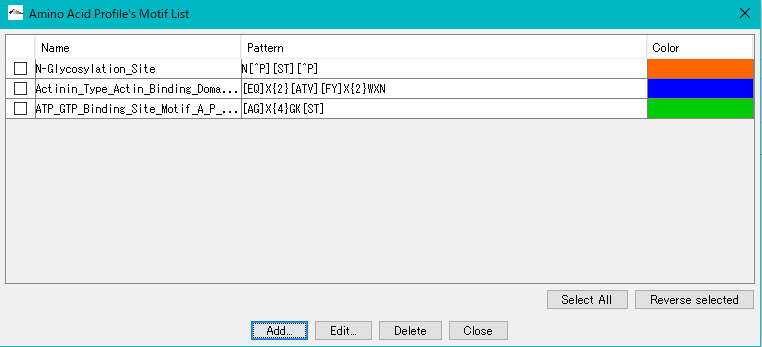
An amino acid sequence motif can be registered in the list.
Each line of the list consists of the following items.
Check box: Checked items will be processed by operation buttons at the bottom.
Name field: The name of the amino acid sequence motif.
Pattern field: Alignment pattern of amino acid sequence motif is described by regular expression.
Color field: Displays the amino acid sequence in the color specified here if the motif is detected on the amino acid sequence lane at the top of the amino acid profile viewer.
Select All button: Click to check all Qualifiers.
Reverse Selected button: When clicked, the checked qualifiers are unchecked and checked for unqualified Qualifiers.
Add ... button: Displays a dialog to add a new motif.
Edit ... button: Displays the edit dialog for editing the selected motif.
Delete button: Deletes the selected motif (multiple processing possible)
Close button: Close Amino Acid Profile's Motif List.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings