IMC T01B Assembly using in silico Assembler
in silico Assembler is de novo Assembler.
Assemble DNA fragments of 50 bp or more.
Reads from NGS can also be assembled (up to about 1 million reads. For larger size use Velvet etc).
In the preprocessing, limited number of processing, designation of minimum QV, maximum N bases can be limited.
Operation
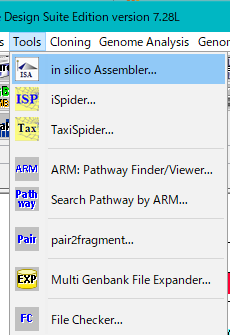
- Select Tools -> in silico Assembler from the menu.
- If using it for the first time, sample data is not installed. You will be asked if you want to install the sample data.
- The in silico Assembler button tool will be displayed.
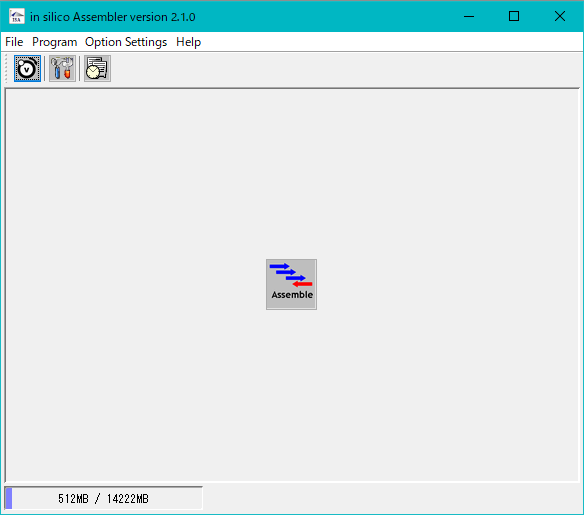
- Click Assemble.
- The Assembly execution dialog is displayed.
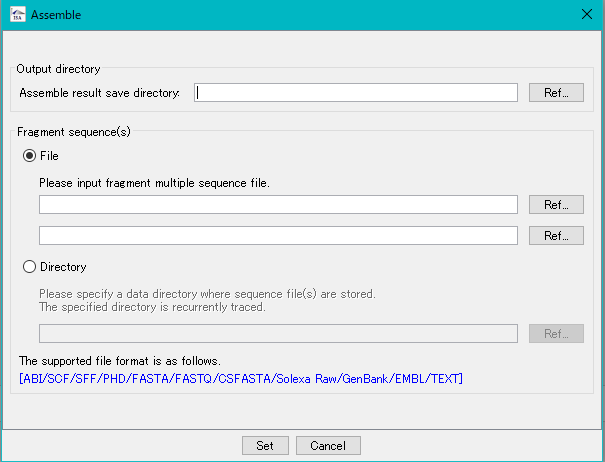
- Specify the save directory of the result.
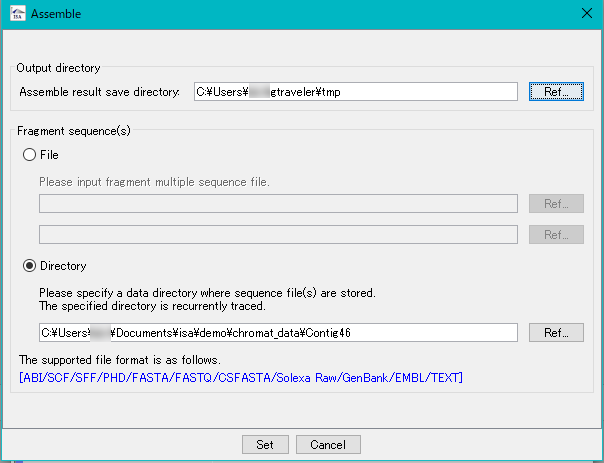
- Specify the DNA fragment files to be assembled.
- The fragment files to be input is specified in one of two ways.
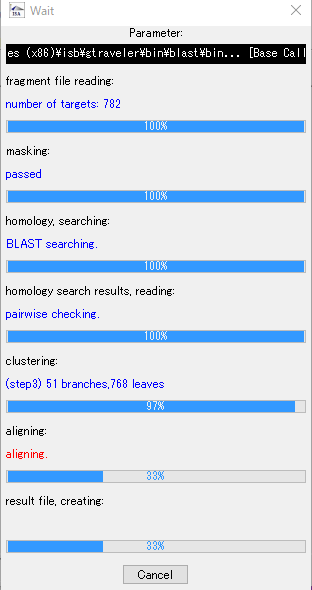
- During assembly execution, a progress message is displayed, so you can grasp the progress of the assembly.
- When complete, a completion message will be displayed.
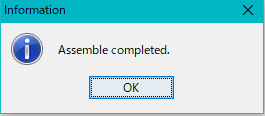
- Click OK.
- The completion message closes.
Points to be noted
The assembly result is stored in the result storage directory specified before execution.
There is a method of mechanically combining a large number of contig sequences generated as assembly results into one sequence before loading.
Assembly results are usually saved as multiple Contig files, so loading as they are divided into a number of fragment files and loaded.
If there are many fragments, there is a method of specifying all of them and not directly loading, generating and loading a multiple GenBank format file mechanically combining them.
To load assembly results into the main feature map, use IMC's normal sequence loading method.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings