IMC 014 Find Homology in Related Genomes
In many situations, homology analysis can be performed easily in the state where the main feature map of one genome and the reference feature map of the closely related species genome are displayed. If sample data is loaded, you can try running homology analysis as an example.
- Click NC_000964.gbk in the Bacillus subtilis directory of the main directory tree.
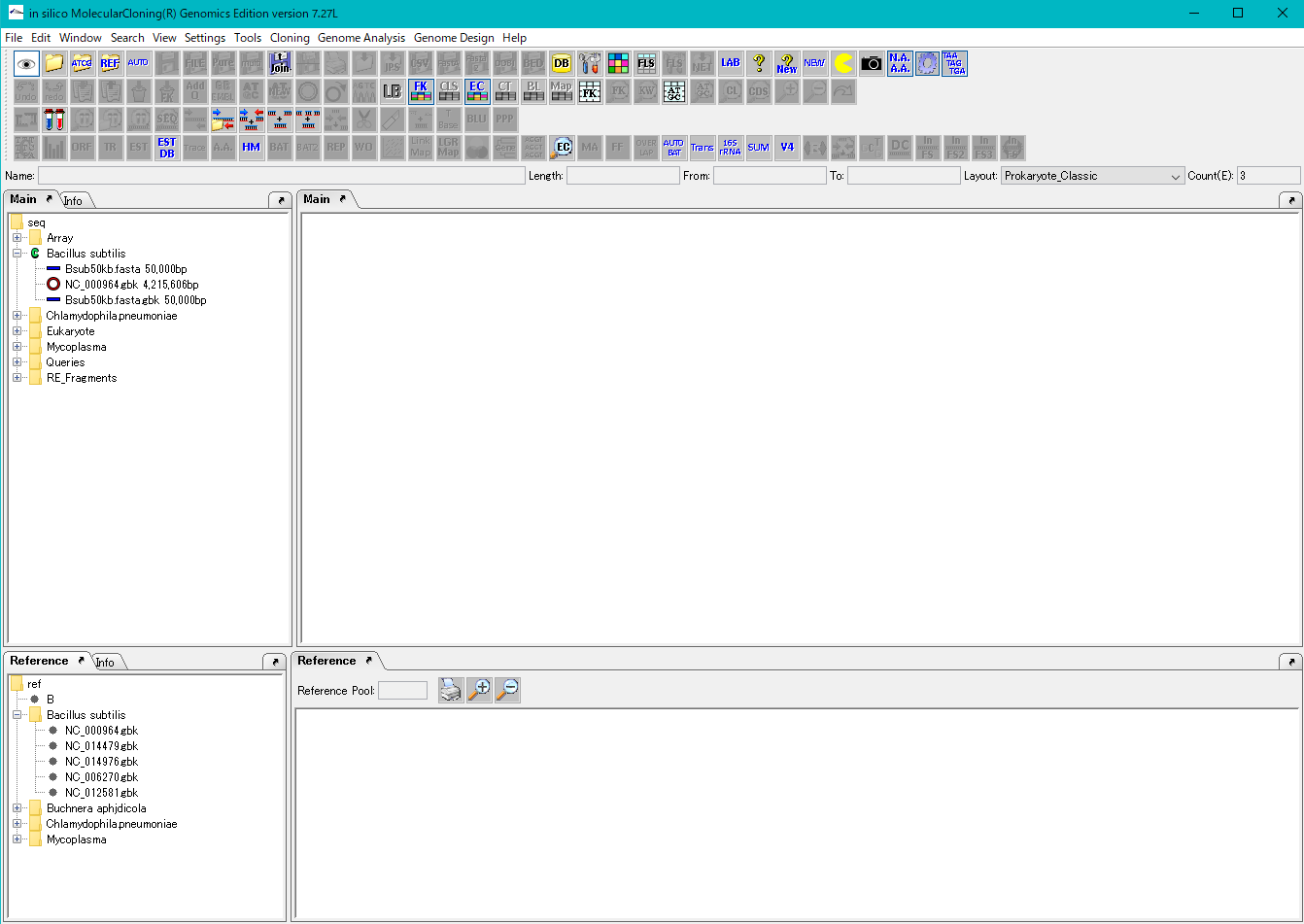
- The background of the array node is red and the map of that genome is displayed in the main feature map.
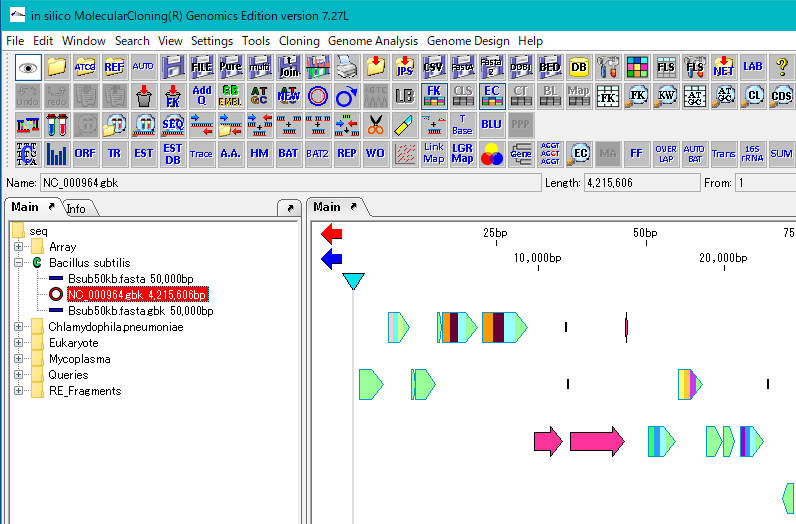
- Right click on the Bacillus subtilis directory in the Reference directory tree to make it the current directory. If the directory icon is a green C mark, it is the current directory.
- In the Reference feature map, maps of two closely related genomes are displayed.
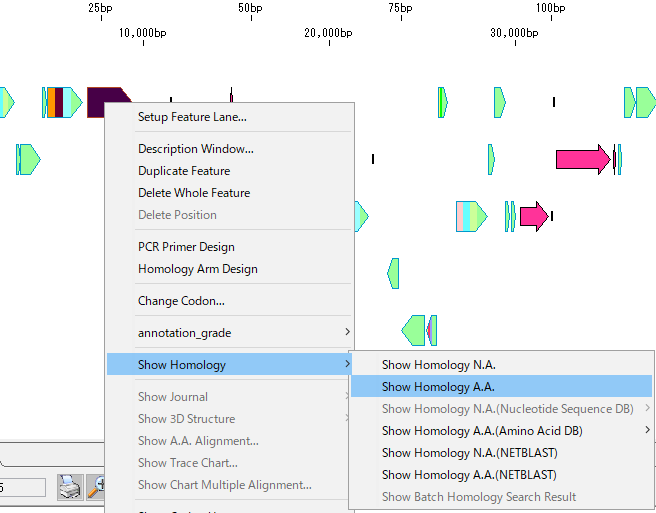
- Right-click on one CDS feature on the main feature map. By default CDS features are displayed with icons of lying houses.
- Popup menu is displayed.
- From the menu, select Show Homology -> Show Homology A.A.
- An execution confirmation message is displayed.
- Click "Yes".
- During execution, progress message is displayed.
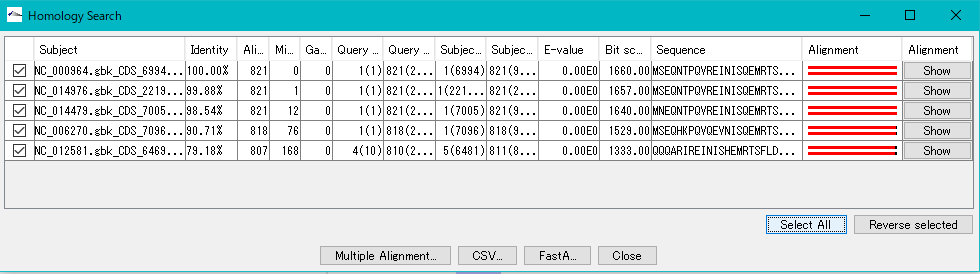
- The Homology Search result dialog is displayed.
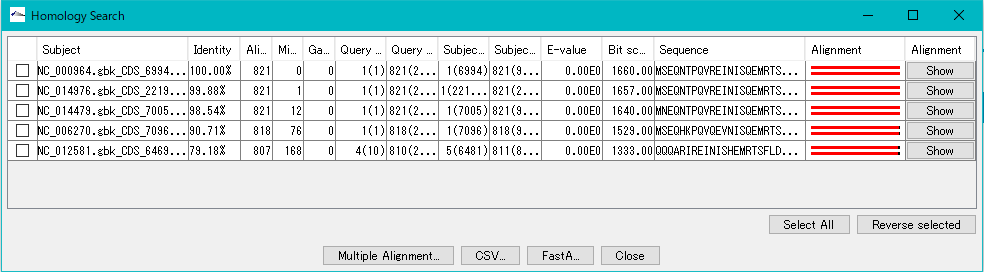
- Click one line.
- The homology genes of the source CDS of the main feature map and the reference feature map are aligned.
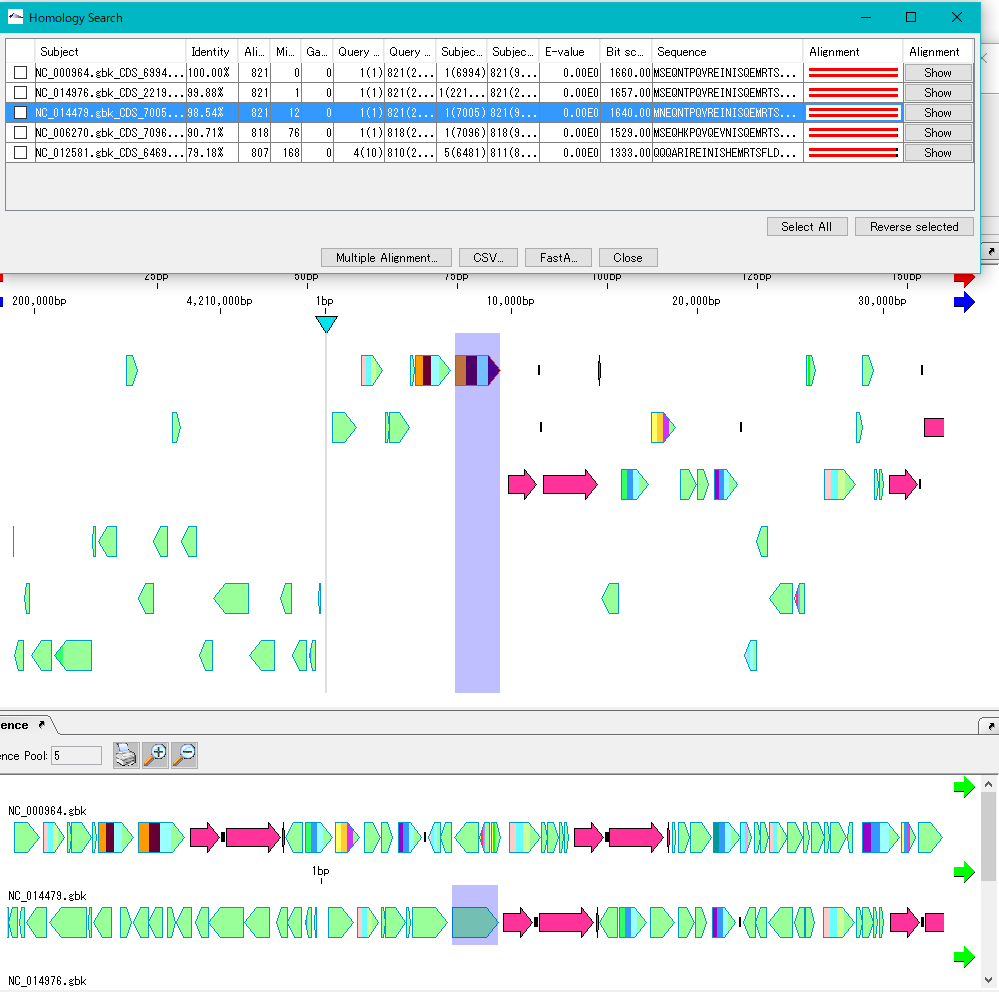
- To see multiple alignment, click Select All and click Multiple Alignment.
- An execution confirmation message is displayed.
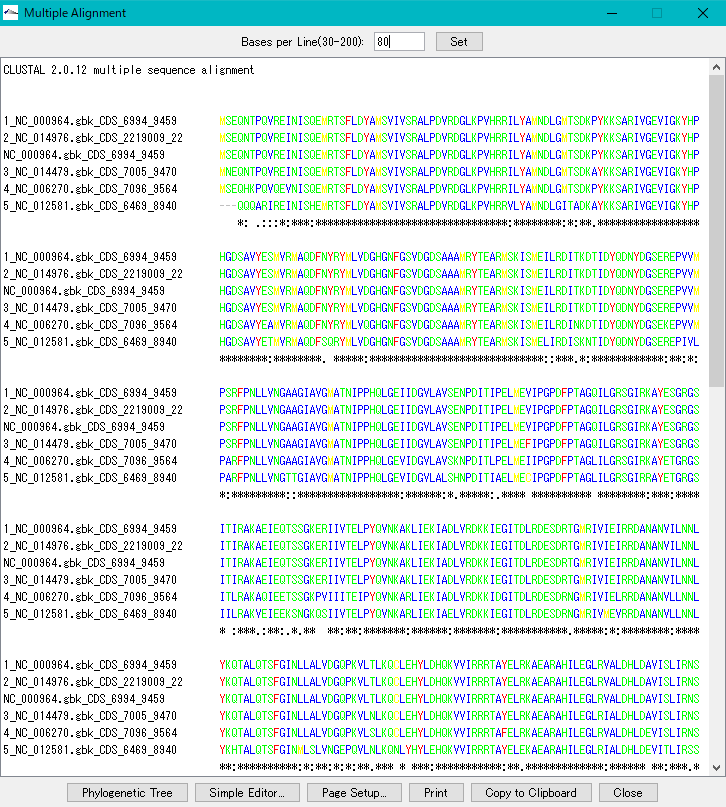
- Multiple alignment is displayed.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings