Registration operation
- Load the template base sequence into the main current directory.
- The loaded sequence is displayed on the main feature map.
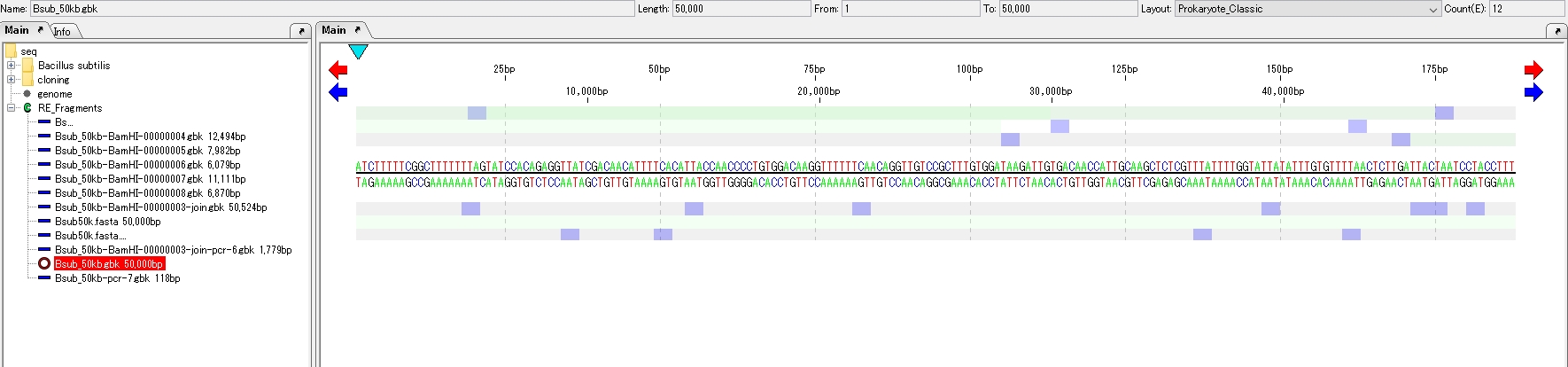
- If the array lane is not displayed in the main feature map, add the array lane to the main feature map.
- Drag the base sequence on the Plus Strand upstream of the region you want to amplify with the mouse.
- The dragged base sequence is highlighted in red.

- Right click the mouse on the nucleotide sequence.
- A pop-up menu will be displayed.
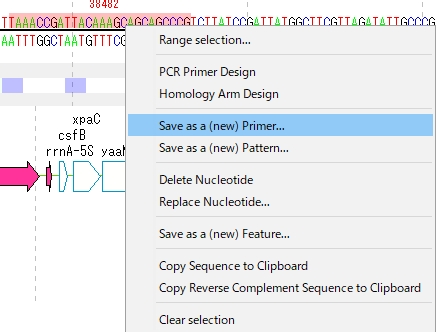
- Click "Save as a Primer" from the menu.
- The “Primer Registration” dialog is displayed.
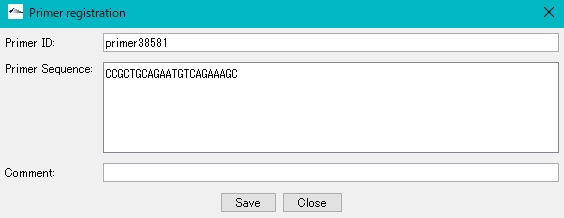
- Enter a unique name in "Primer ID" (primer + base position number is set by default).
- In "Primer Sequence", the copied base sequence is automatically pasted in uppercase letters.
- You can write freely in the "Comment" field.
- Click "Save".
- The Forward primer is registered.
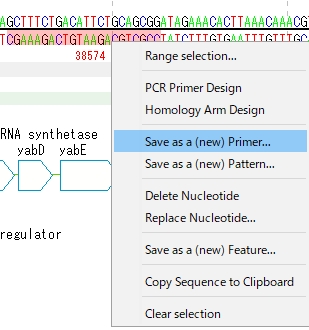
- Register the Reverse Primer in the same way.
- In the case of Reverse primer, drag Minus Strand II downstream of the PCR amplification region in the sequence lane with a mouse.
-
Checking registered primers
- Click Cloning-> Primer Design-> PCR Primer Registration from the menu.
- The Priming Site Search Window will be displayed.
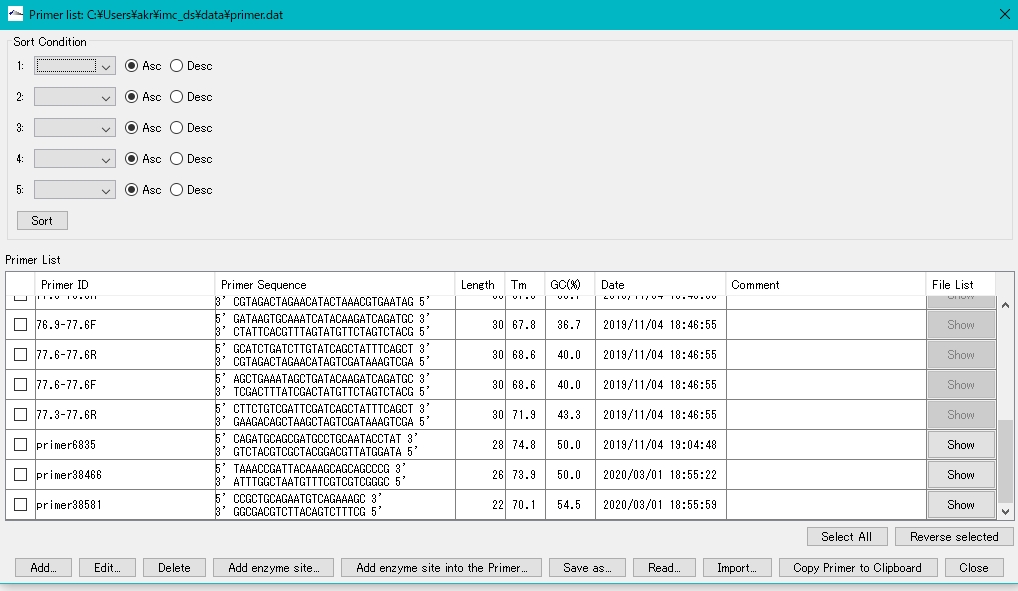
- Here, the registered primers are displayed along with their sequence, Tm, and GC content.
- Recently registered primers are at the bottom of the list.






 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings