Operation
Area designation
- Drag any area on the feature map with the mouse.
- The dragged area is displayed in red.
Primer design
- Click the right mouse button on the feature map.
- A pop-up menu will be displayed.
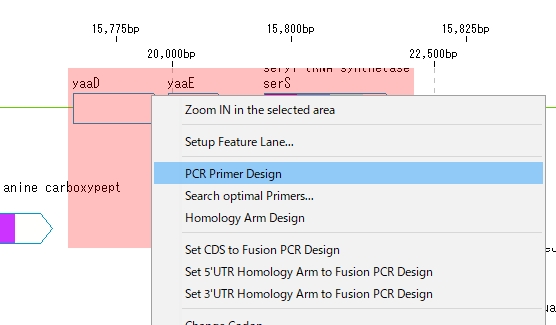
- Select "PCR Primer Design" from the menu.
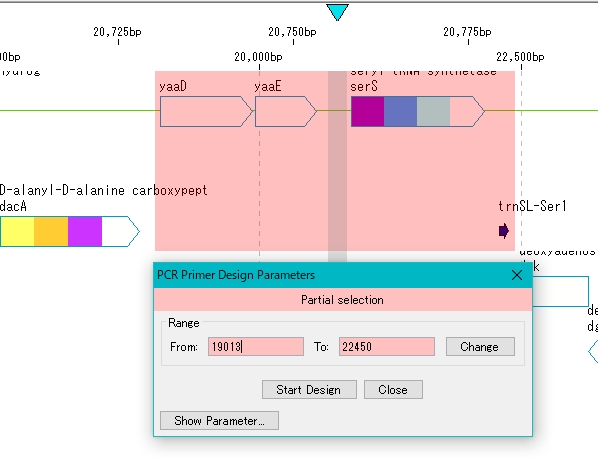
- The “PCR Primer Design” dialog is displayed.
When setting different from the default
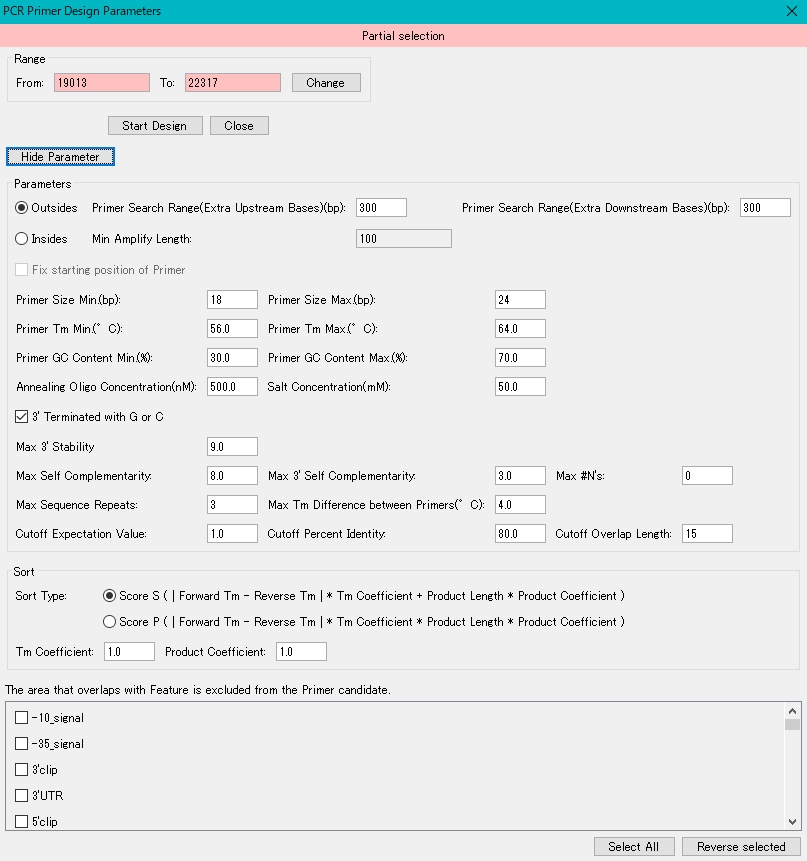
- Click Show Parameters in the PCR Primer Design dialog.
- The detailed parameters are displayed.
- To change the region to be amplified, enter a value directly in the "From:" and "To:" fields, and click the "Change" button.
- The amplification target area is changed.
- To change how many bases upstream of the amplification region are searched for the primer priming site, enter a number directly in the “Primer Search Range (Extra Upstream Bases) (bp)” field.
- In this case, the primer is searched from the base position of From: up to the number of bases entered in this field.
- Set the same for the downstream side.
- For the range of the base length of the primer, enter the minimum base length and the maximum base length directly in the “Primer Size Range Max.” Field.
- For the Tm range of the primer, enter the minimum Tm temperature (° C) and maximum Tm temperature (° C) directly in the “Tm Range Min.” And “Tm Range Max.” Fields.
- For the range of the GC content of the primer, enter the minimum GC content (%) and the maximum GC content (%) directly in the "GC Range Min." And "GC Range Max." Fields.
- To specify the annealing oligo concentration, enter the concentration (nM) directly in the "Annealing Oligo Concentration" field.
- To specify the salt concentration, enter the circular concentration (nM) directly in the "Salt Concentration" field.
- Indicates the score threshold for judging whether there is primer self-complementarity.
- If the score is less than or equal to the Max SelfarComplementarity field score, the primer is acceptable.
- When detecting repetitive sequences in the primers, specify the maximum base length that will not be recognized as a repetitive sequence in the "Max Sequence Repeats" field.
- That is, if there are two or more identical sequences with a base length longer than this base length, they will not be used as primers.
- Specify the maximum Tm difference between primer pairs by directly entering it in the "Max Tm Differenece" field.
- Three parameters are prepared to check whether there is a priming site in the target DNA region.
- For the check, Blast1 is used, so the parameters are the same as those of Blast.
- “Cutoff Expectation Value” indicates the expected value that is the detection threshold.
- "Cutoff Percent Identity" indicates the threshold value of what percentage or more of the primer base lengths match.
- “Cutoff Overlap Length” indicates the minimum number of base overlaps between the primer and the priming DNA sequence.
- Click the "Set" button.
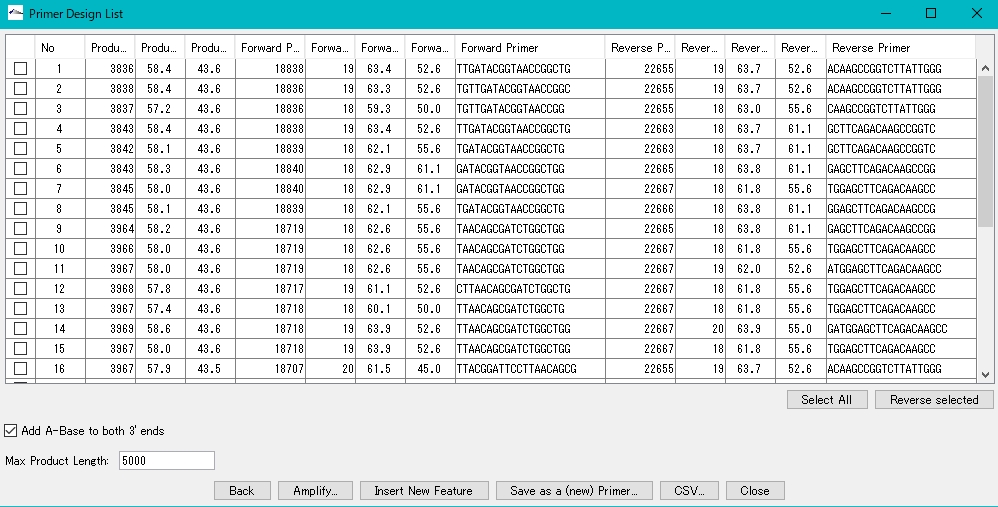
- A window listing the optimal PCR Primer Set appears.
- An error message is displayed if the optimal Primer Set cannot be designed.
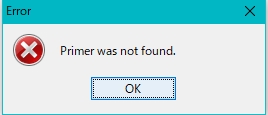
- If you cannot design the Primer, change the design parameters and redesign.
Confirmation of amplification region
- Clicking on each line of the Primer Set List displays the amplification region corresponding to that Primer Set in color.
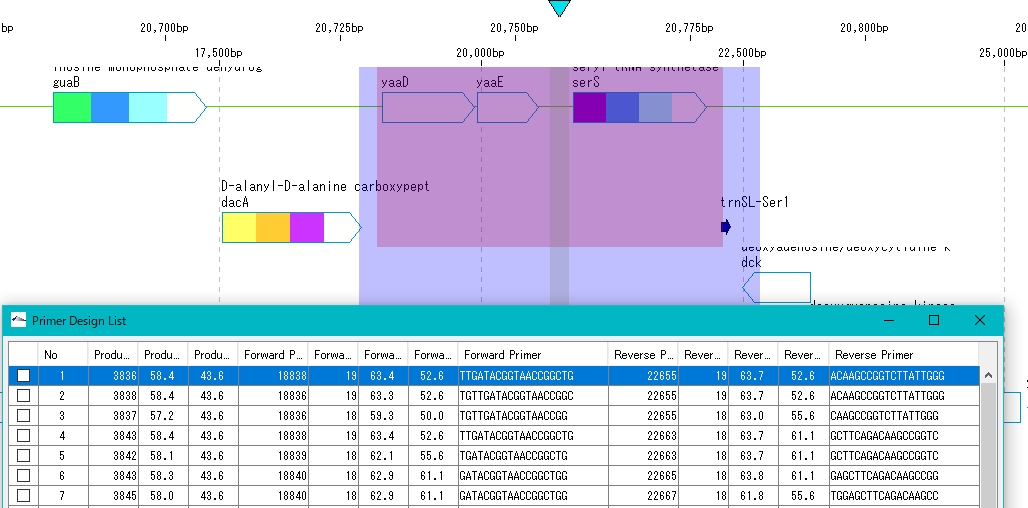
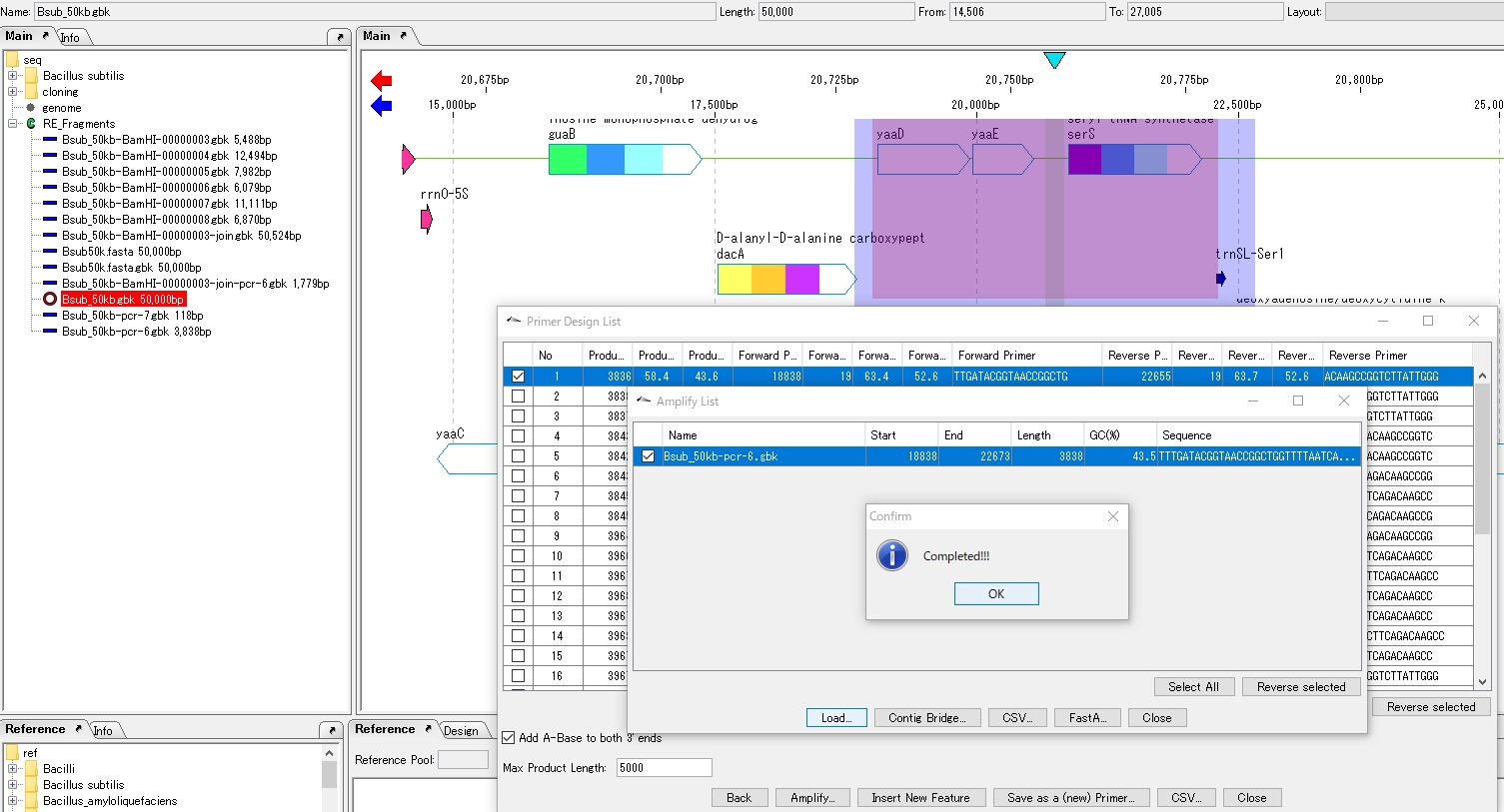
Perform PCR amplification
- Check the box of Primer Set to be used from Primer Design {List}.
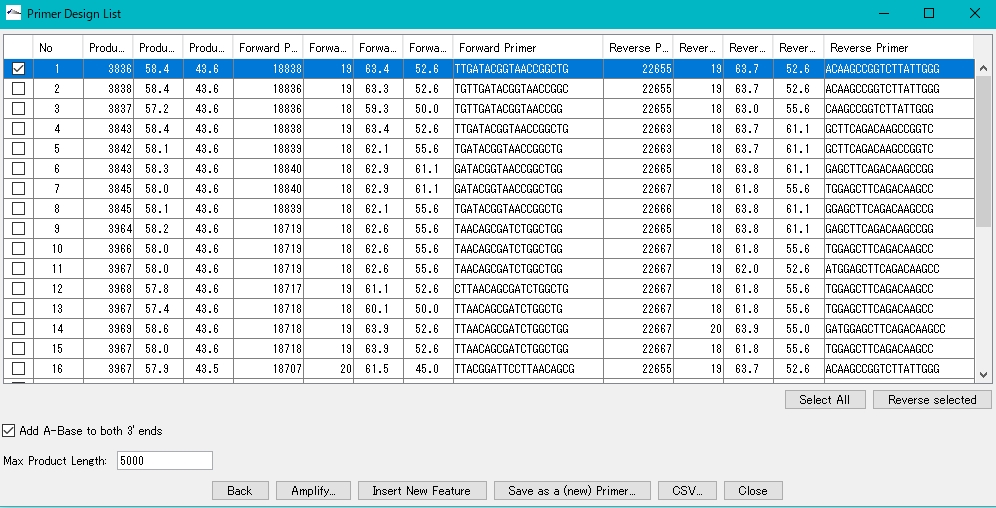
- Click Amplify.
- The confirmation message “Start PCR” appears.
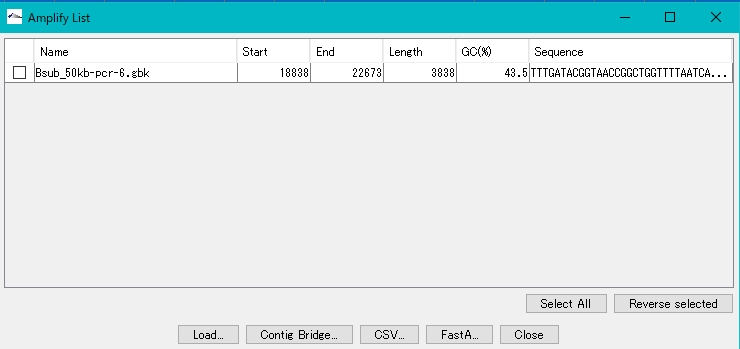
- The Amplify List window appears.The list lists the amplification products.
Loading amplification products
- To add an amplification product to the main directory, check the checkbox for that product.
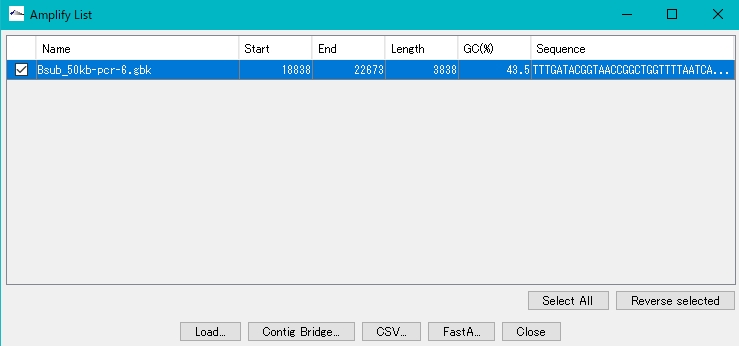
- Click Load.
- The Load is executed, and when it is completed, a completion message is displayed.
- The generated sequence of the PCR product is added to the current main directory.
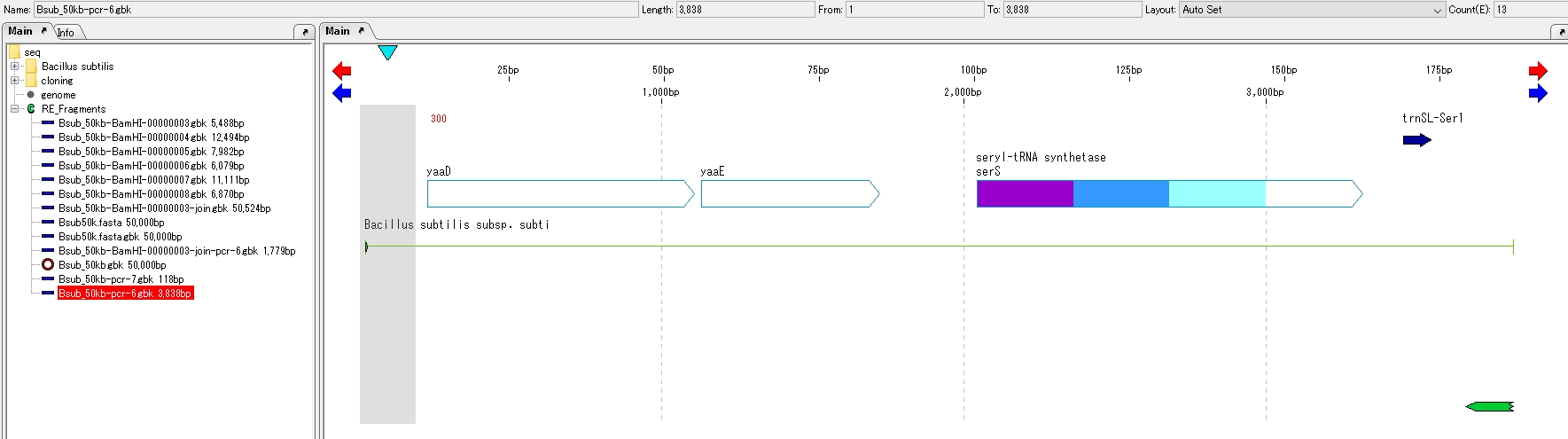
Browse amplification product sequences
- Click the sequence of the PCR product added to the current main directory.
- The feature map of the PCR product is displayed.














 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings