IMC F05 Functions of Circular Genome Map Viewer
Circular Genome Map (Circular Genome Feature Map) is a function to compare and draw Feature and Content from arbitrary circular chromosomes or circular chromosomes of closely related species on a concentric circle and arbitrary numerical data specific to genome position.
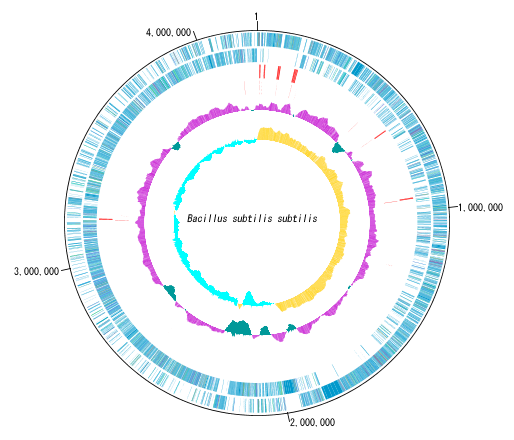
You can display distribution status of each gene such as genes of each circular chromosome, change of base composition, etc.
The order of concentric circles of Feature and Content to be placed can be changed arbitrarily.
Drawing parameter setting can be changed for each concentric circle.
It is also possible to change the diameter of the circular genome map and the printing parameters.
Printing a circular genome map In addition to direct printing, you can output files in PDF, PNG, and EMF formats. In case
A large number of features drawn on the concentric circle of the circular genome map can be classified and displayed in different colors depending on the annotation type if there are annotations. In case
- Circular Feature Lane Different Feature drawing independent color setting is possible.
- Feature Key Draw with different colors.
- When Color Qualifier is set, draw the CDS Feature with the specified color.
- Draw the CDS Feature with the color specified in the Classification code.
- Draw the CDS Feature with color derived from EC Nubmer.
- Draw the CDS Feature with the color specified in the Category code.
- Draw the CDS Feature with the color specified in Blast's score range.
Array files drawn on the circular genome map are referenced in the order shown in the main current directory.
For this reason it is easy to manipulate drawing a number of different circular genome sequences in the same format.
By clicking on a part of the circular genome map it is possible to display the area of the corresponding main feature map.
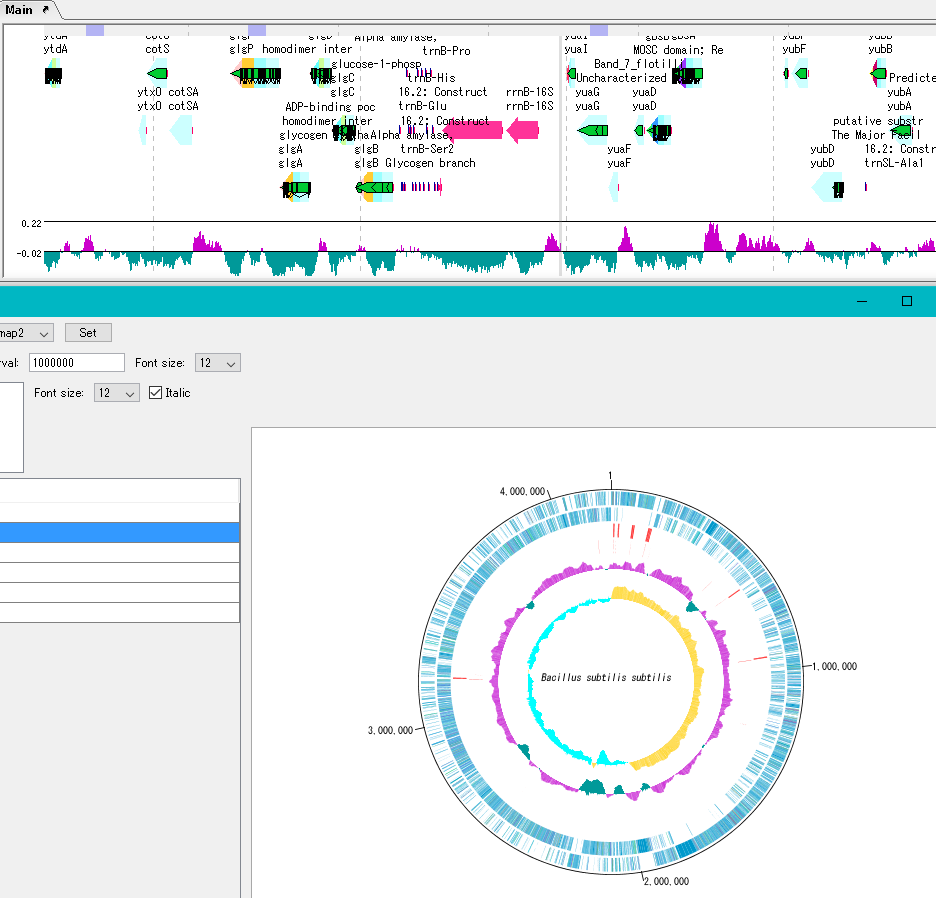
In the circular genome map, the sequence file name and total base length (when single chromosome) are drawn at the center, and the base number scale is displayed around it.
The starting position base of the scale can also be changed.
The map layout can be registered as a layout style, and the registered circular genome layout style can be called later.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings