IMC W212C Semi Auto Annotation Dialog from Reference Genome
Auto Annotation by Reference Data result dialog for operation.
- Add annotations semi-automatically to the main current genome sequence with the reference genome as the database for Blast search.
- With this annotation function you can annotate multiple genome sequences at once, so the Name field will display a list of annotated main current genome sequences.
- If you click on one of them, the results belonging to that array are displayed.
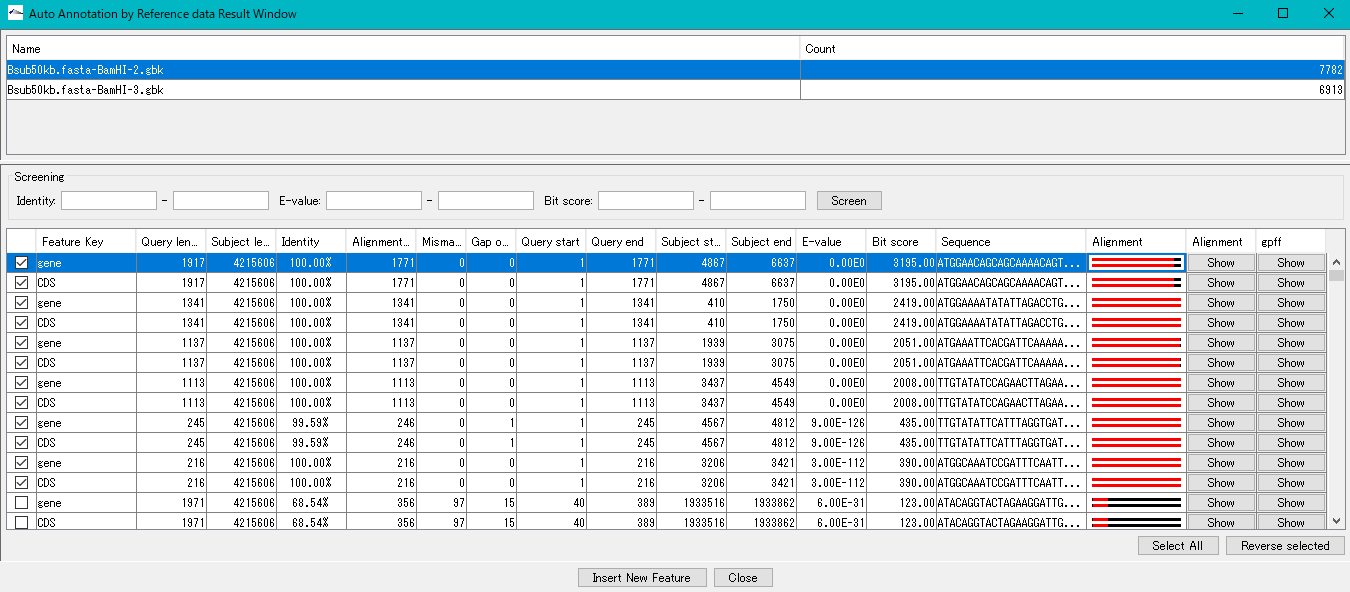
The Screening field is for narrowing down search results. Three key types are as follows.
- Identity: The degree of agreement (percentage) in the alignment region between the query sequence and the hit sequence
- E-Value: The probability that a sequence that coincidentally coincides in this search space when compared with a random sequence of the same composition
- Bit Score: Search Sum of match / mismatch score of query array and hit array specified in matrix to be used Screen: Enter the range of each value and click this button to narrow down.
Hit list: Hit subjects are listed for each feature of the reference genome as a query sequence.
Explanation by column
- Check box: Selected when checked Feature Key: Feature key of the query feature on the reference genome
- Query Length: Base length of the feature to be a query on the reference genome
- Subject Length: Base length of reference genome sequence
- Identity: The degree of agreement (percentage) in the alignment region between the query sequence and the hit sequence
- Alignment Length: Base length of alignment region
- Mismatches: Mismatching number of bases between query sequence and hit subject sequence in alignment region
- Gap Openings: the base length of the generated gap in the alignment region
- Query Start: Relative starting base position of the query feature sequence
- Query End: Relative end base position of query feature sequence
- Subject Start: Absolute starting base position of reference genome sequence
- Subject End: absolute end base position of reference genome sequence
- E-Value: The probability that a sequence that coincidentally coincides in this search space when compared with a random sequence of the same composition
- Bit Score: Search Sum of match / mismatch score of query array and hit array specified in matrix to be used
- Sequence: Array of alignment regions of hit subjects
- Alignment (Figure): An illustration of the total length and alignment region of the query sequence and the hit subject, respectively
- Alignment (Button): When clicked, a pairwise alignment dialog of query array and hit subject array is displayed.
- Gpff (Button): When clicked, the Feature Key, Qualifier, Value list of the GPFF of the hit subject is displayed, and the selected entry can be copied as Qualifier.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings