IMC B2DA Operations Performed from Selected Area of Feature Lane
- By dragging the mouse on the feature lane of the main feature map you can select that area.
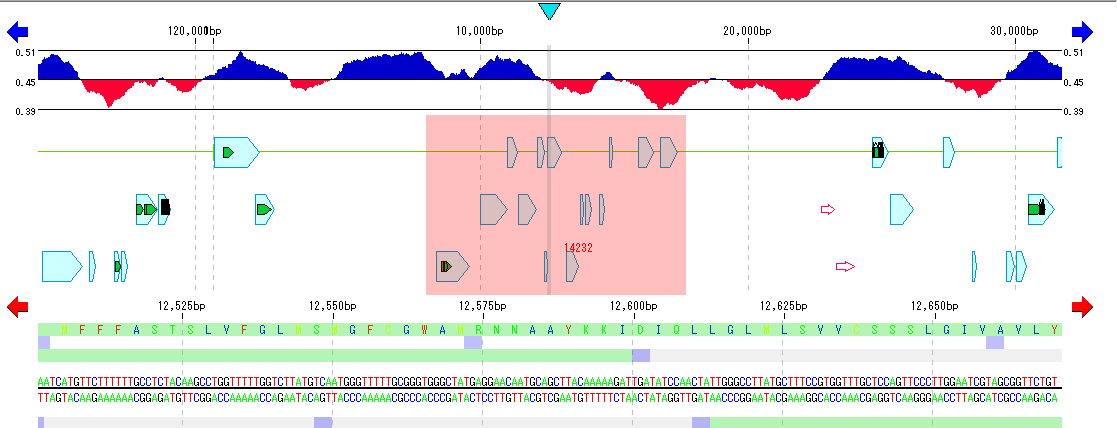
- The following functions can be executed by right-clicking the mouse on the selected area.
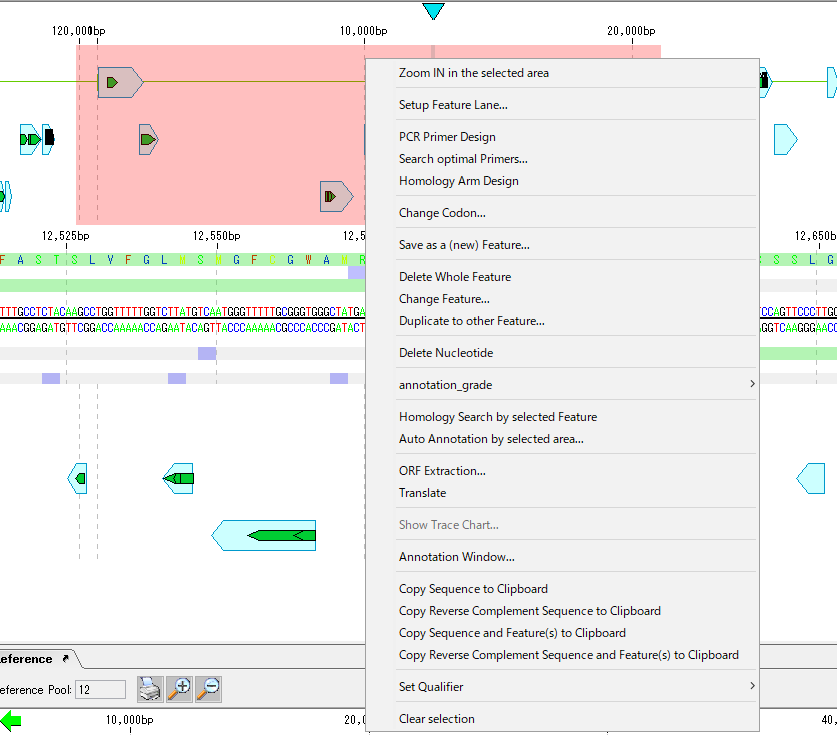
- Zoom IN the Selected Area: Zoom in the selected area to the entire screen
- Set Feature Lane: Set feature lane
- PCR Primer Design: Perform PCR primer design to amplify the selected region
- Search Optimal Primers: Search registered primers that can amplify selected regions
- Design Homology Arm: Design a homology arm with a selected region as a homologous recombination region
- Replace Codon: Codon substitution of the designated restriction enzyme recognition site in the selected region, eliminating the restriction enzyme recognition site
- Save as a (New) Feature: Registers the selected area as a new feature
- Delete Whole Feature: Deletes features in selected area
- Change Feature Key: Change the feature key of the feature in the selection area
- Duplicate Features: Register replicas of features included in selection area with designated feature key
- Join Features: Join features contained in selection area and register as new features
- Annotation Grade: Sets the annotation_grade of the feature in the selected area
- Homology Search by Selceted Features: Performs a Blast search with the features contained in the selection area as the query sequence
- Translate: Translate CDS / ORF features included in selection area into amino acids
- Show Trace Chart: Displays the trace waveforms included in the selected area
- Delete Nucleotide: Deletes the base sequence of the selected region
- Auto Annotation by Selected Area ...: Automatic annotation of selection area
- ORF Extraction: Extracts ORF from selection area
- Launch Annotation Window: Launches an annotation window whose selection range is the operation range
- Copy Sequence to Clipboard: Copies the base sequence of the selection area to the clipboard
- Copy Reverse Complement Sequence to Clipboard: Copies the reverse complementary base sequence of the selected region to the clipboard
- Copy Sequence and Features to Clipboard: Copies the nucleotide sequence of the selected region and features contained in the region to the clipboard
- Copy Reverse Complement Sequence and Features to Clipboard: Reverse complementary base sequence of selected region and region Copy the included features to the clipboard
- Set Qualifier: Specify the Qualifier of the feature included in the selection area at once
- / color
- / label
- / featuremap
- / genomemap
- / trans_table
- Clear Selection: Deselects the selection area
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings