- Perform a homology search with COG, which is an ortholog database, on the genomic base sequence for which CDS has been identified, and automatically post the COG function classification code. As a result, each CDS can be displayed in color classification on the feature map.
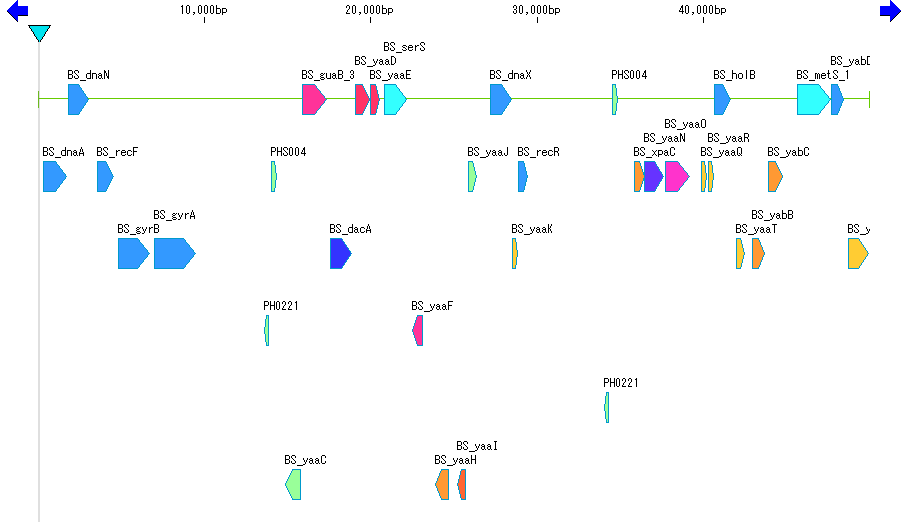
Implemented Edition
Preparation
- COG database myva
- Genomic sequence file: for example Bsub 50 kb.gbk
- Although it can be executed with any genomic sequence, the eukaryotic genome sequence is not suitable as the COG database does not contain prokaryotic species other than yeast.
- The sample sequence is taken from a partial region of B. subtilis, but if it is executed in the whole genome, processing time may be required.
Operation
- Download the COG database.
- It can be downloaded from NCBI or from our site download menu.
- Register the COG database.
- Use the DB registration function for batch homology search.
- Select "File" -> "Create Blast DB" in Menu. Alternatively, click Create Blast DB in the toolbox.
- Detailed information on "Create Blast DB" function
- Load genome sequence.
- The sample genomic sequence is generated in the "Users / Documents / sample" folder during IMC installation.
- Or you can download from our download menu.
- Execute ORF Extraction.
- Select "Genome Analysis" -> "ORF" -> "ORF Extraction ..." from the menu.
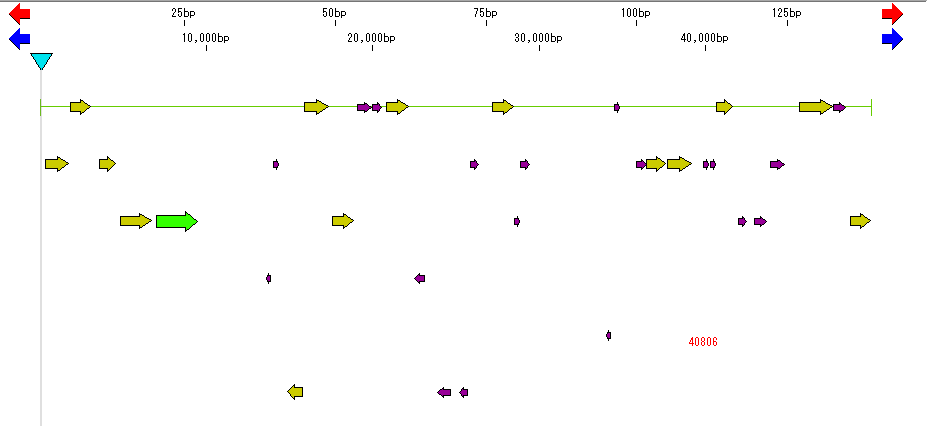
- Perform amino acid translation, convert each ORF candidate to CDS, and at the same time perform that amino acid translation.
- Select "Genome Analysis" -> "ORF" -> "Translation" from the menu.
- The Translation Run dialog is displayed.
- If no amino acid sequence is registered in each CDS, the search will be "No Hit".
- This process is unnecessary if the amino acid sequence has already been registered in each CDS.
- Each ORF candidate is converted to CDS and amino acid translated using the specified Genetic Table.
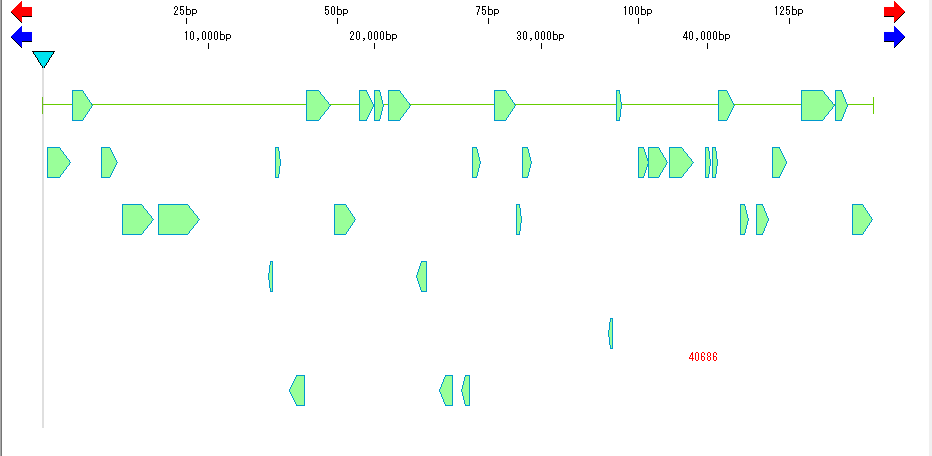
- Detailed information on amino acid translation function of CDS
- Perform batch homology search.
- To target all CDSs on the genome, select "Genome Analysis" -> "Homology Search" -> "Homology Search for Selected Feature Key" in the Menu or click the Bat Homology Search button in the tool box.
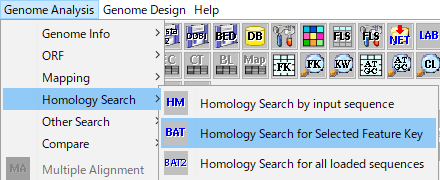
- If you want to target all CDSs in a specified area on the genome, drag the area with the mouse and specify the area. Right-click on the selection area and select "Homology Search by
- Selected Feature" from the displayed pop-up menu.
- In the Batch Homology Search dialog that appears, select the COG database and click the Set button to start execution.
- Detailed information on Batch Homology Search function
- Classification code is displayed by Classification Code.
- Click the "Draw Feature in Classification Color" button on Toolbox.


 , AE
, AE , DS
, DS , GT
, GT





 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings