IMC W441 Ligation Dialog Description
In the Ligation dialog, a list of the DNA sequences loaded in the main current directory is displayed and ligation of the selected sequence is executed.
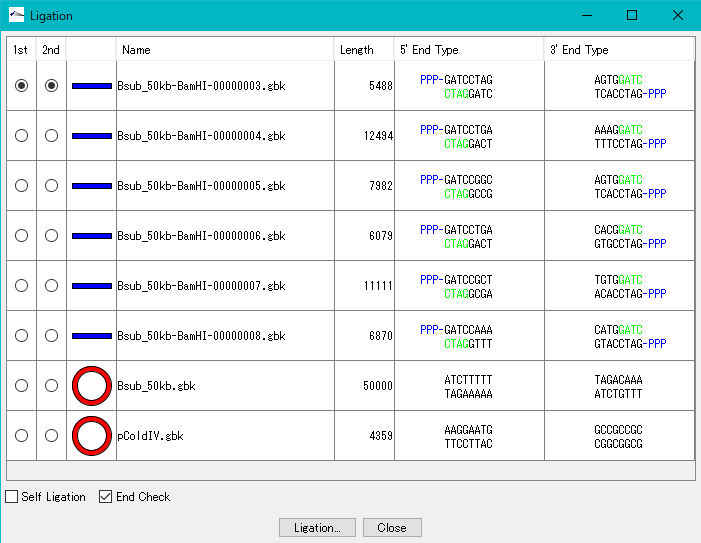
1st radio button: When on, we make that sequence the first fragment.
2nd radio button: When on, sets the sequence as the second fragment.
DNA shape field:  is displayed when the array is circular,
is displayed when the array is circular,  is displayed in the case of Linear.
is displayed in the case of Linear.
Name field: The file name of the DNA sequence file is displayed.
Length field: The sequence length of the DNA base sequence is displayed.
5 'End Type field: The 5' end shape of the DNA base sequence is displayed. If phosphorylated, PPP- is added. The bases displayed in green indicate base pairs that do not exist. That is, it indicates that the base shown in black protrudes with a single strand.
3 'End Type field: The 3' end shape of the DNA base sequence is displayed. If phosphorylated, PPP- is added. The bases displayed in green indicate base pairs that do not exist. That is, it indicates that the base shown in black protrudes with a single strand.
Self Ligation check box: If checked, the ligation to be executed will be self-ligation. The 2nd radio button column becomes Inactive. When unchecked, it becomes a ligation reaction between two sequences. 2nd radio button becomes active.
End Check checkbox: If checked, ligation will be performed if the terminal shape is complementary and phosphorylated. Ends that do not fit are not subject to ligation.
Ligation execution button: When this button is clicked, ligation judgment between the selected DNA sequences is executed.
Close button: Click this button to close the dialog without executing ligation.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings