about macOS 11 Big Sur
Although macOS 11 Big Sur has been released, the following problems have been reported regarding our products when the version is upgraded to the OS.
1. The dongle does not work properly. The Corresponding Dongle Driver has not released from the maker yet.
The workaround is either of the following.
- Revert to macOS 10.
- Switch from dongle license to activation license (free of charge). Please contact us.
IMC B2K1 Register CDS Candidate from Frame Lane
- CDS candidates can be registered as new features from the stop lane stop absence region of the frame lane.
- This is not a CDS prediction function. The GT-AG rule is ignored.
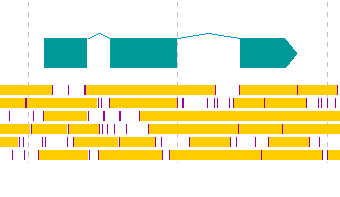
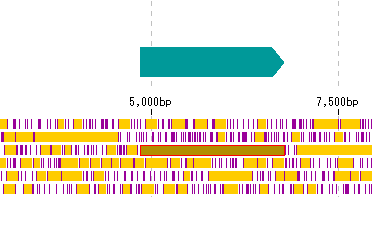
Operation
YD0018 New Feature Registration Bug from Frame Lane
Phenomenon
In Frame Lane, if you select a region where the stop codon does not appear and register that region as a new feature by right-clicking the mouse, that region will be complemented in the Description Window even though it is in Forward Strand.However, it occurs in the case of new registration from the 3rd frame of Forward Strand. Registration from the 1st and 2nd frames works normally.
Immediate countermeasures
Uncheck Complement in the Description Window and register.
Counter-measure
Bug fix Version IMC Ver.7.46 has been released (2020/12/09).
YD0017 License is not recognized while the dongle is lit
Phenomenon
- The LED of the dongle is lit, but the license is not recognized, the "License request" dialog is displayed, and IMC cannot be started.
Cause
- Due to the recent Java Version Up, activation may not work properly.
Coping
- Reverts the Java version to the previous version.
- If you cannot change the Java version to the previous version, please contact in silico biology, inc. Temporarily issue a fixed license.
IMC T07A Expand Multiple GenBank File (GBFF) into Taxonomy Tree
Extract the GBFF file consisting of many single GanBank format files into a tree-like directory according to the Taxonomy description of each GenBank file and place it in IMC so that it can be handled easily.
Background
The sequence data of GenBank and RefSeq are compiled into a large compressed file at each release time.
Each entry consists of a header file (*. Gbff) and an array part (* fna). In order to make a single GenBank file, it is necessary to combine these two kinds of files.
GenBank File Expander decompresses these files of GenBank and RefSeq, synthesizes them, and unfolds them as a phylogenetic tree-like directory in an arbitrary place.
If you specify this expanded directory as IMC's root directory, it can be displayed and loaded in IMC's directory tree.
Operation
Click "Tools -> Multiple GenBank File Expander" from the menu.
IMC S411 Execute OGAB Fragmentation Designer
To rebuild the designed gene cluster, fragment the cluster optimally.
After each fragment is chemically synthesized, each fragment is reconstituted by OGAB method to prepare a long chain DNA cluster.
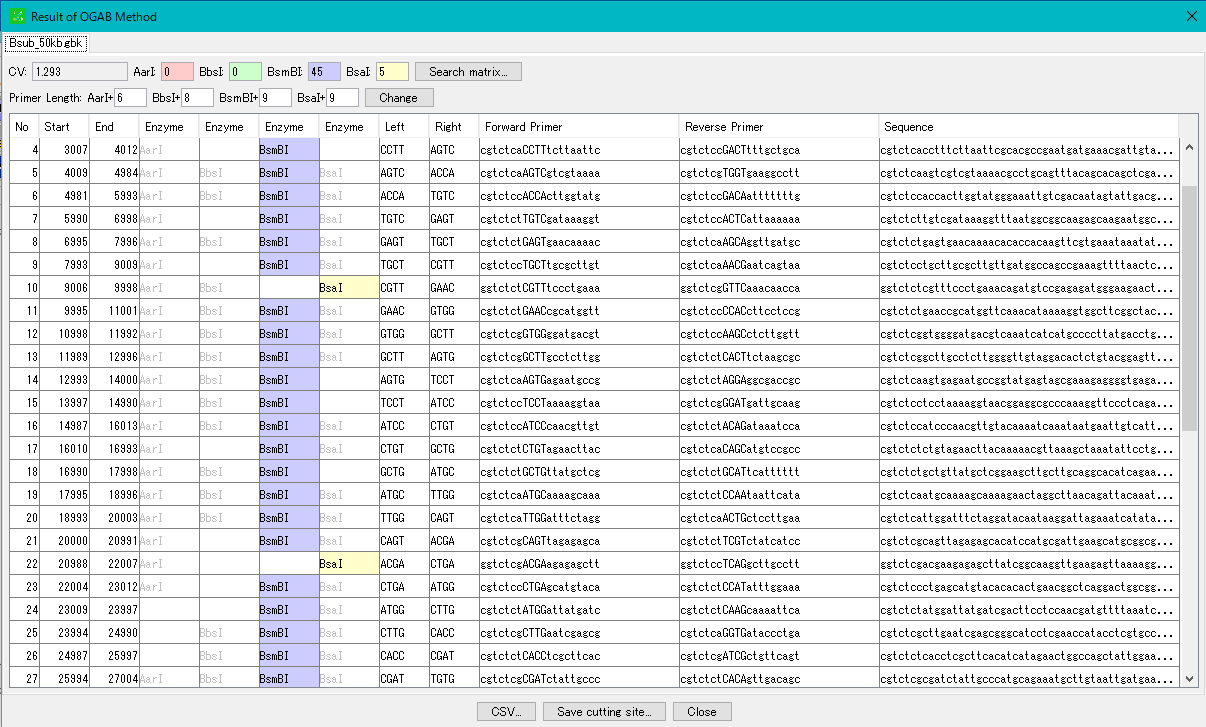
Implementation Edition
This function is implemented in the following edition.
IMCDS![]()
Operation
IMC O11A Operation of Core Genome Analysis
Extract the core genome from a pan genome.
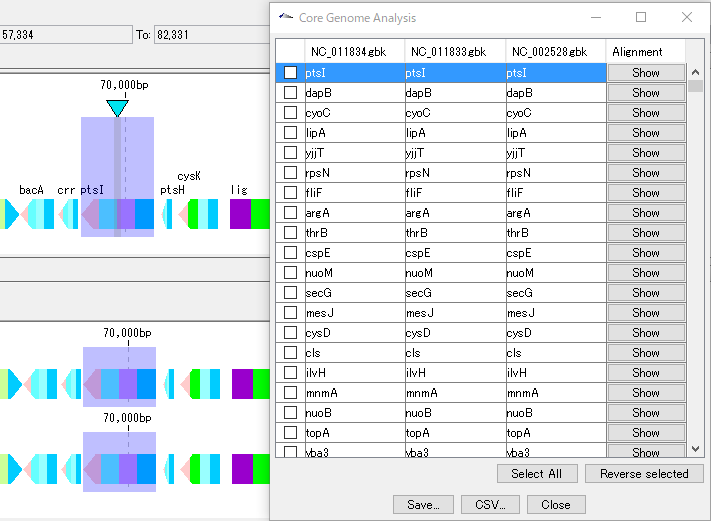
Preparation
Load one genome into the current main feature map.
Load multiple genomes to compare to the current reference feature map.
IMC O13A Operation of Mutation Search
Detects one base mutation between closely related genomes and generates a list of all mutations. In addition, from the list you can display the variation of the reference feature map and the base position of the corresponding main feature map.
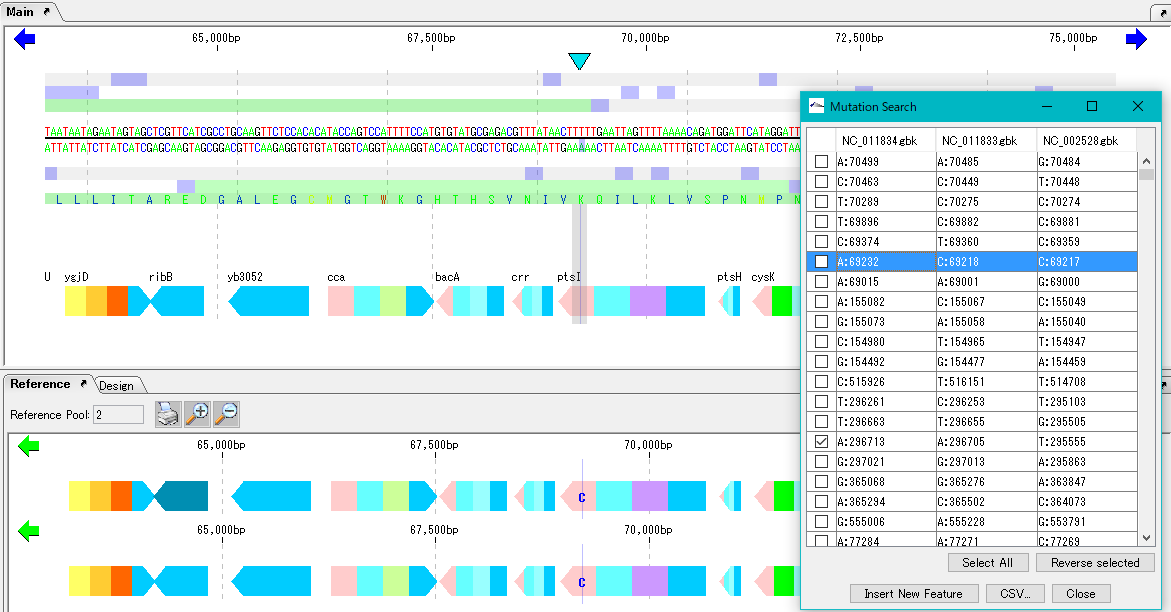
Preparation
Load the base stock genome into the current main feature map.
Load multiple closely related genomes to compare with the current reference feature map.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings