IMC B2DA41 ORF / CDS of Selected Region on Feature Lane is Translated into Amino Acid
The CDS features and ORF candidate features included in the selected area on the feature lane of the main feature map are translated into amino acids.
The ORF candidate feature automatically changes the feature key to CDS.
You can change the genetic code table used for amino acid translation.
Operation
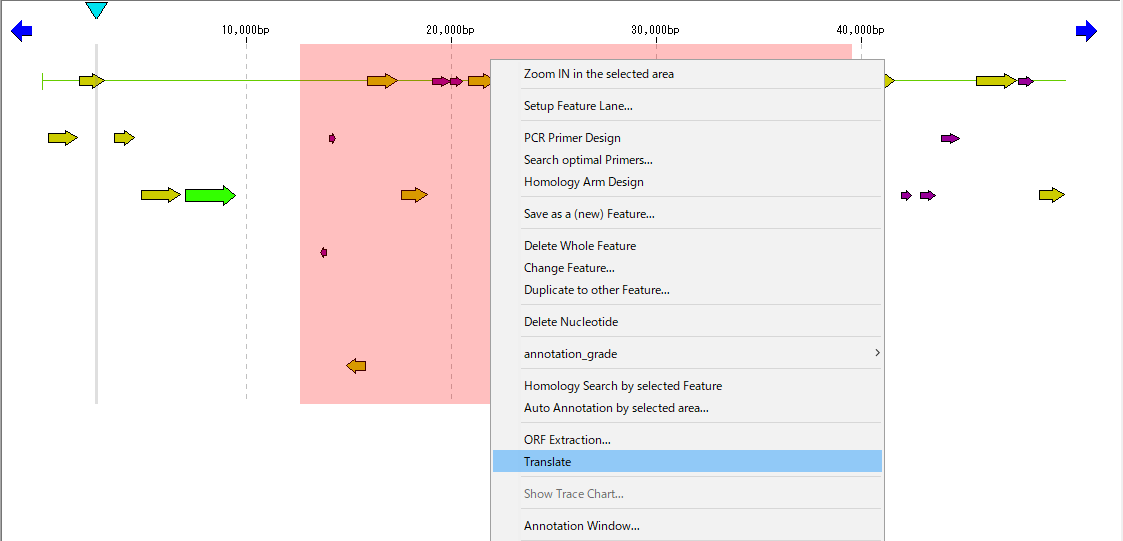
- Right-click on the selection area of the feature lane of the main feature map.
- The menu will be displayed.
- Select "Translate".
- The "Translation Setting" dialog will be displayed.
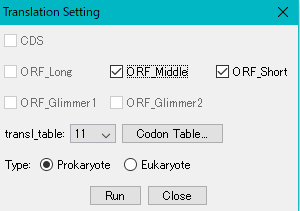
- Only the CDS / ORF feature included in the selection area can be checked.
- If you need to change the genetic code table, select that number from the "Transl_table" pull-down menu.
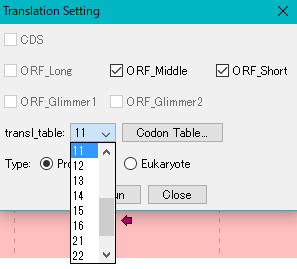
- When clicking "CodonTable" button, the genetic code table corresponding to that number is displayed.
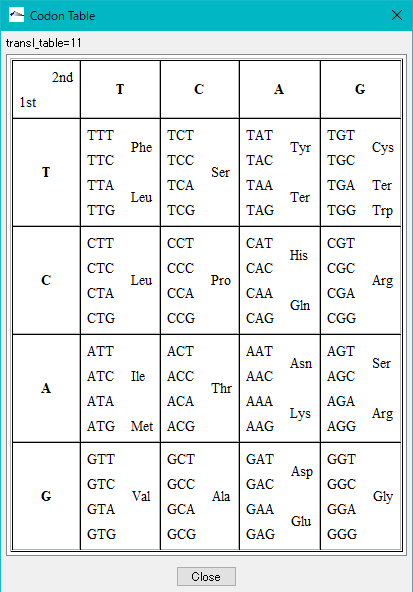
- Click the "Close" button.
- The "Codon Table" is closed.
- Click "Run".
- Each ORF feature is changed to CDS, each amino acid translation is based on the genetic code table.

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings