IMC O04A Draw Venn Diagram
Implemented on
IMC GE![]() AE
AE![]() DS
DS![]()
GT
Function
Using GenBank format files of three closely related genomic nucleotide sequences whose genes have already been identified, it is judged whether or not they are common genes among all the genes present on each genome, Draw a Venn Diagram between 2 genomes.
- For the determination of common genes, amino acid sequence searches by NCBI Blast are used.
- The criterion uses Percent Identity and Overlap Length of the Blast result.
- We define pairs whose Percent Identity and Overlap Length are greater than or equal to the specified value to be common.
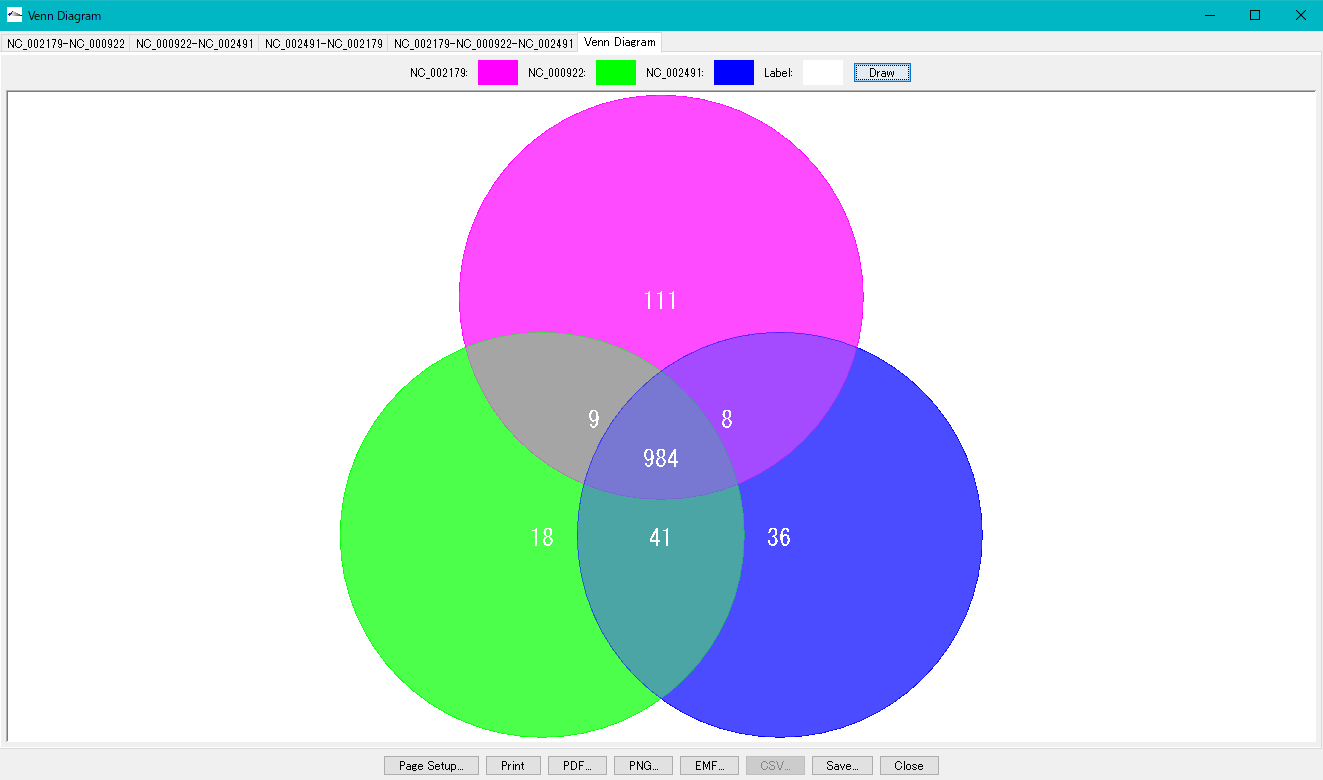
Draw a color graphic on Venn Diagram.
- Click each numerical value to display the gene list.
- It is possible to change the drawing color.
- Printing is possible.
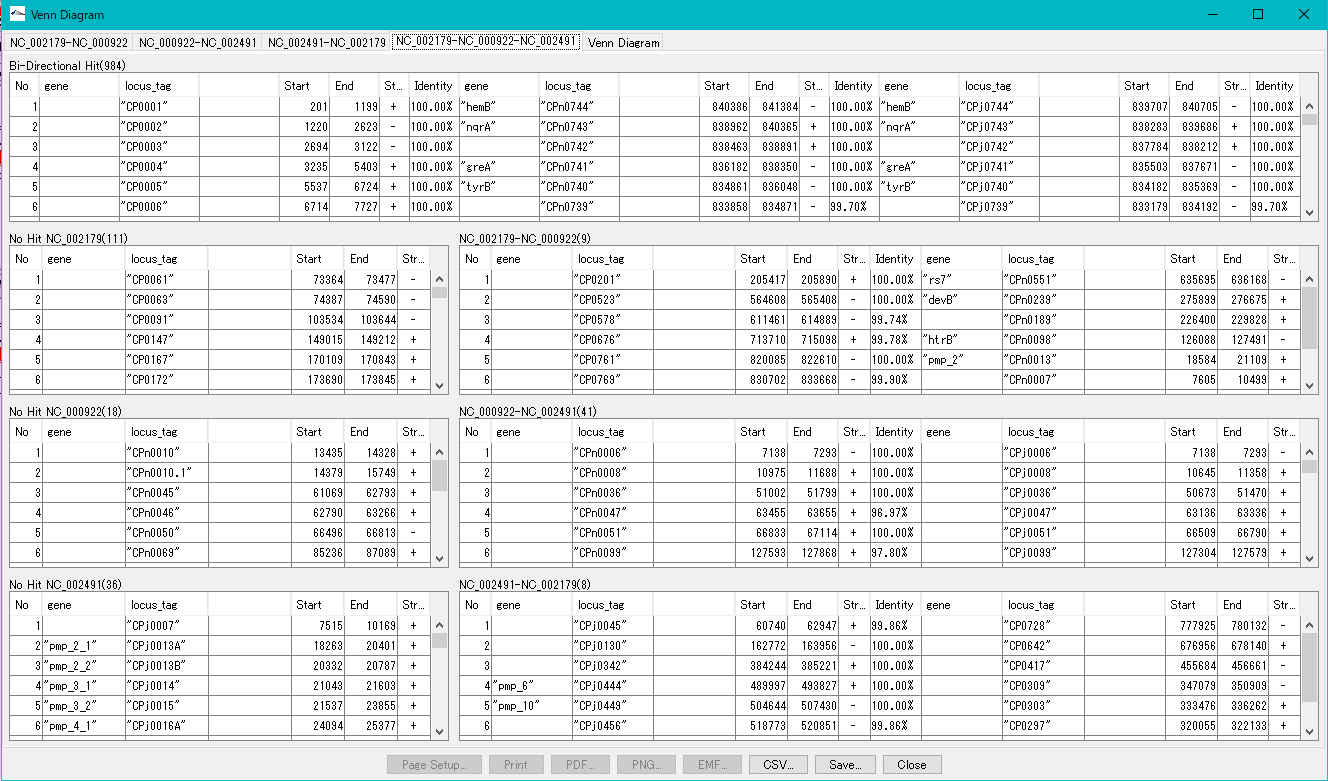
Display a list of common genes etc., output the file.
- Number of genes common to 3 species and their list
- Among the three species, the number of genes common to any two species and their list
- The number of genes that exist only in the genome and genes that do not share genes common to the other two genomes and their list
- Each list can be output as a CSV file.
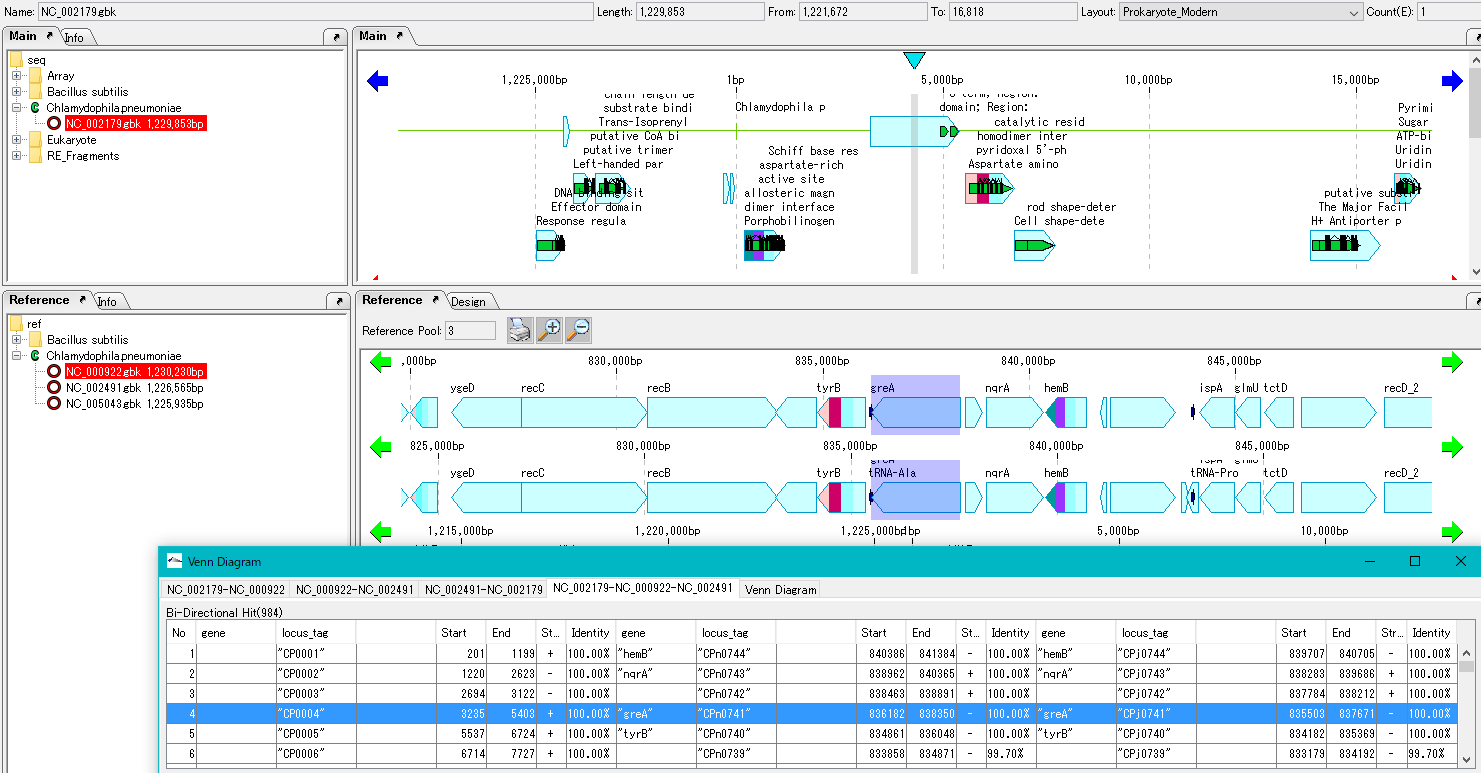
Genetic feature alignment is done for common genes or unique genes.
- By clicking each common gene or unique gene on the list, each genome on MGV (reference genomic map) is genetic feature aligned at that gene position.
Save and Load Results
- You can save the result and load it and view it later.
- If there is no necessary array data in the current directory, the array file is also loaded automatically.
Restrictions
- The Venn Diagram function is implemented in IMCGE / AE / DS. It is not implemented in IMCEE, EE
- All the genome to be drawn by Venn Diagram must be loaded on MGV.
Algorithm
- We compare the amino acid sequence of the CDS identified on the genome to be compared using NCBI Blast's compatibility search.
- In the case of 2 genome comparison, one CDS on one genome performs amino acid sequence compatibility search for all CDS of the other genome, and the top hit CDS matches the query CDS of the original genome In case you define it as a common gene.
- However, if the specified condition (Percent Identity and Overlap Length) is less than the criterion value, it will be rejected.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings