IMC L03A Annotation of Current Genome Sequence by Batch Homology Search
(Batch) homology search to the genome sequence in which ORF extraction and amino acid-translated CDS features are registered and automatically register entry information of the hit database as the Qualifier of the CDS feature.
This function is implemented in the following editions.
IMCGE![]() , IMCPE
, IMCPE![]()
Operation
Load the genome sequence containing the amino acid translated CDS feature into the main current directory.
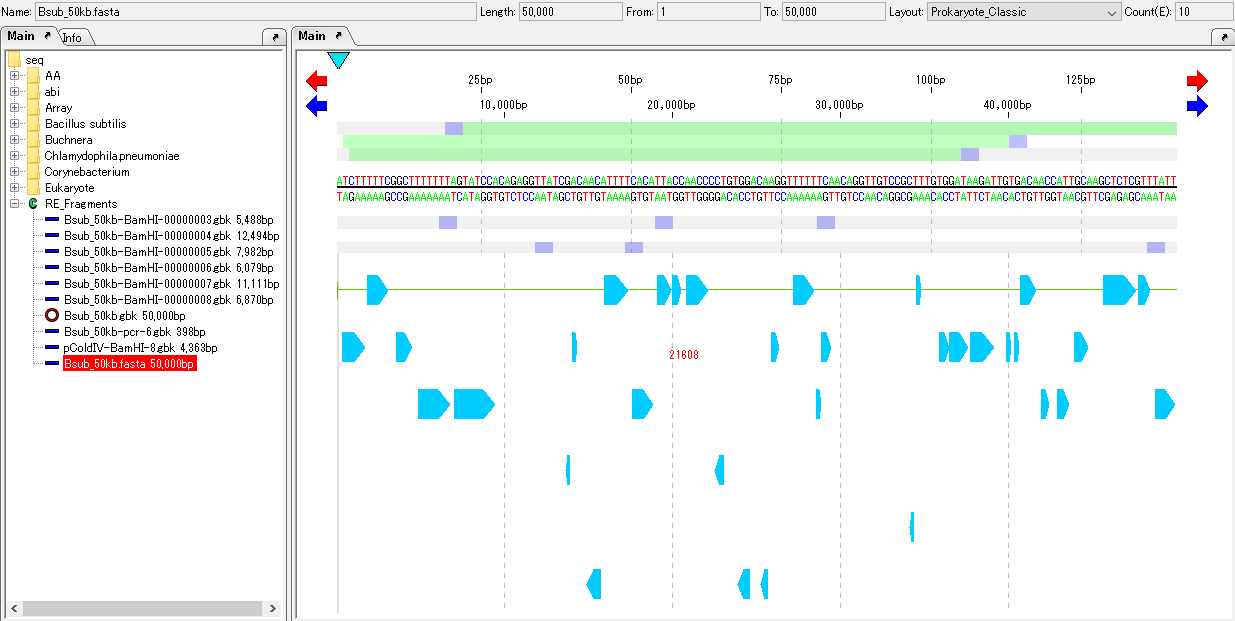
Load GenBank or EMBL format files already annotated relative genome sequences into the current reference map.
Multiple genomic sequences can be loaded.
Please refer here if there is no closely related genome already annotated.
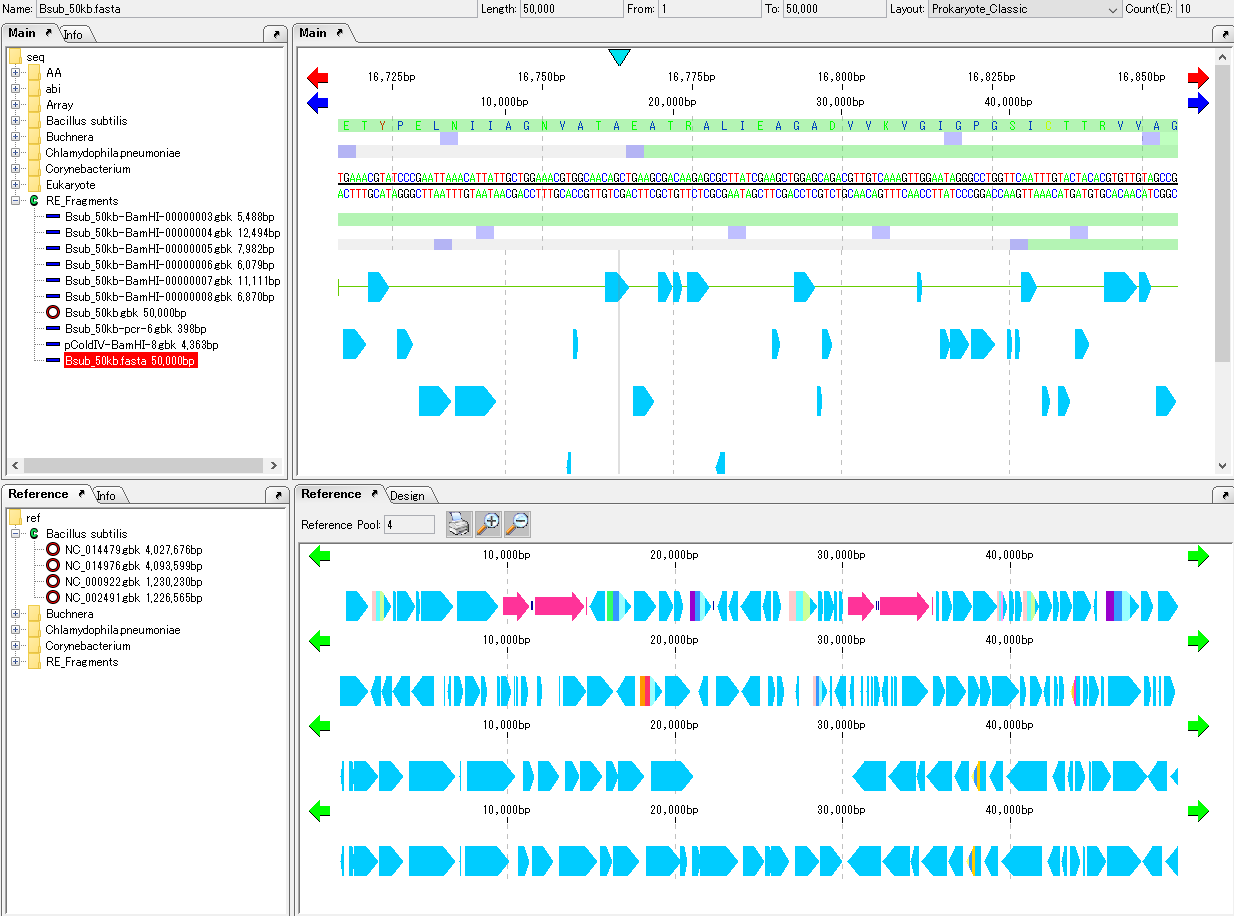
Turn on "Auto Copy" function. For the operation method, please click here.
From the menu click Genome Analysis -> Homology Search -> Homology Search for Selected Feature Key.
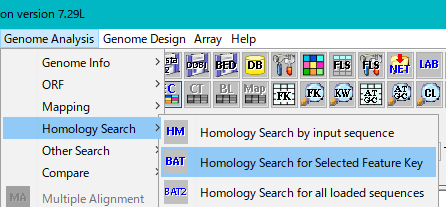
The "Batch Homology Setting" dialog will be displayed.
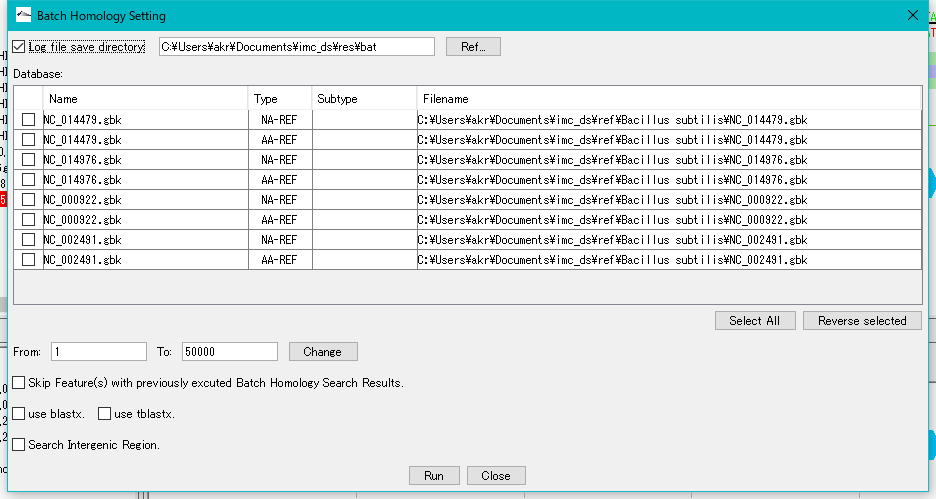
Check "Log File Save Directory" and specify a writable directory.
For each sequence loaded in the current reference directory, databases for Blast search of base sequence and amino acid sequence are automatically generated in the Database list.
Check the amino acid sequence database (AA - REF) of each closely related genome.
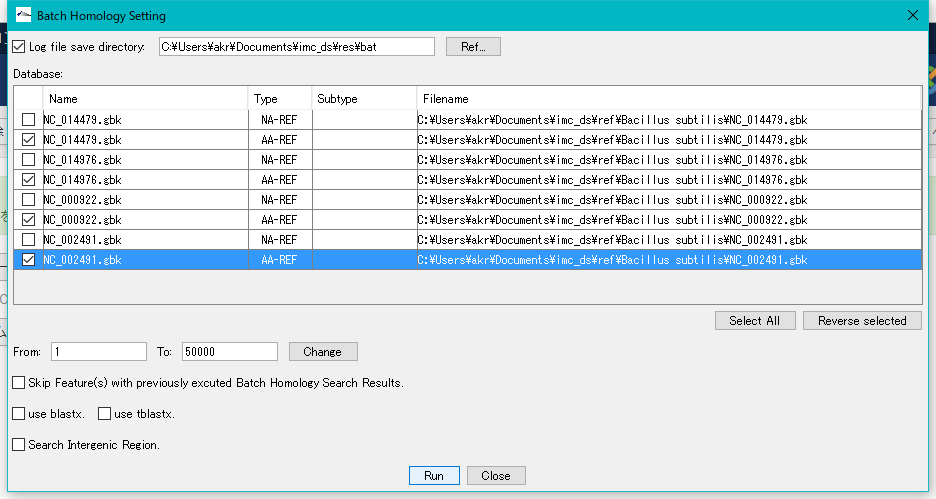
Click "Run".
A confirmation message "Start Homology Search?" Is displayed.
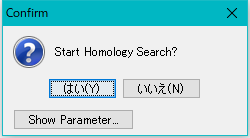
Click "Yes".
Execution of homology search is started, and progress message is displayed during execution.
When the execution is completed, the "Homology Search" result display dialog is displayed.
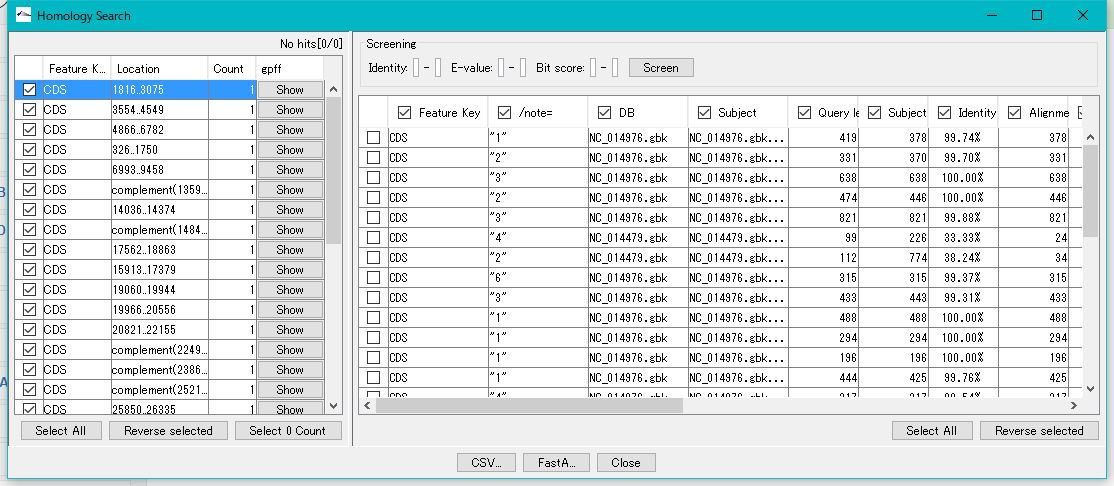
When "Auto Copy" is set, the entry information of the top hit are automatically posted to each CDS feature and added as a comment.
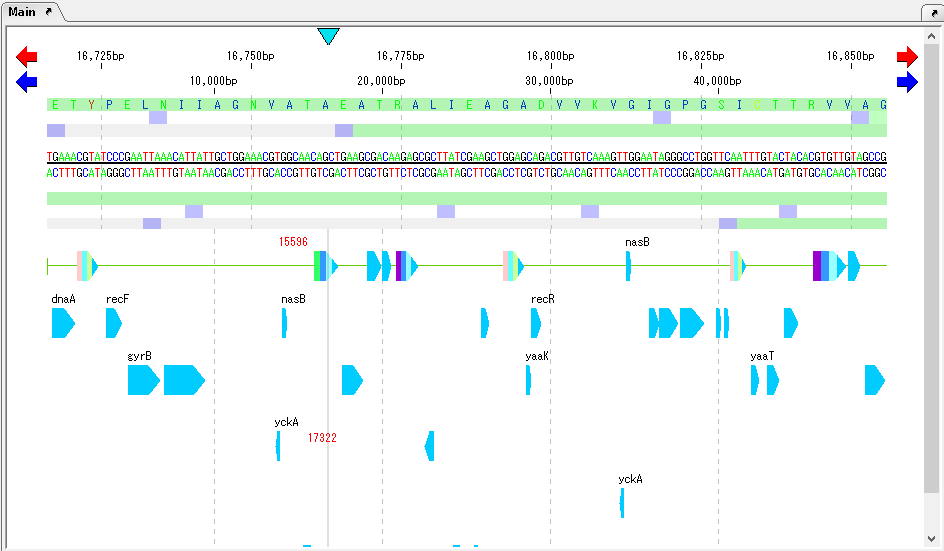
As moving the mouse over one CDS, the Tooltip is displayed and the annotated content is displayed.

 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings