IMC F01 GenBank/EMBL Viewer, Functions and Operations
The GenBank / EMBL viewer is the window that displays the contents of the current main sequence file.
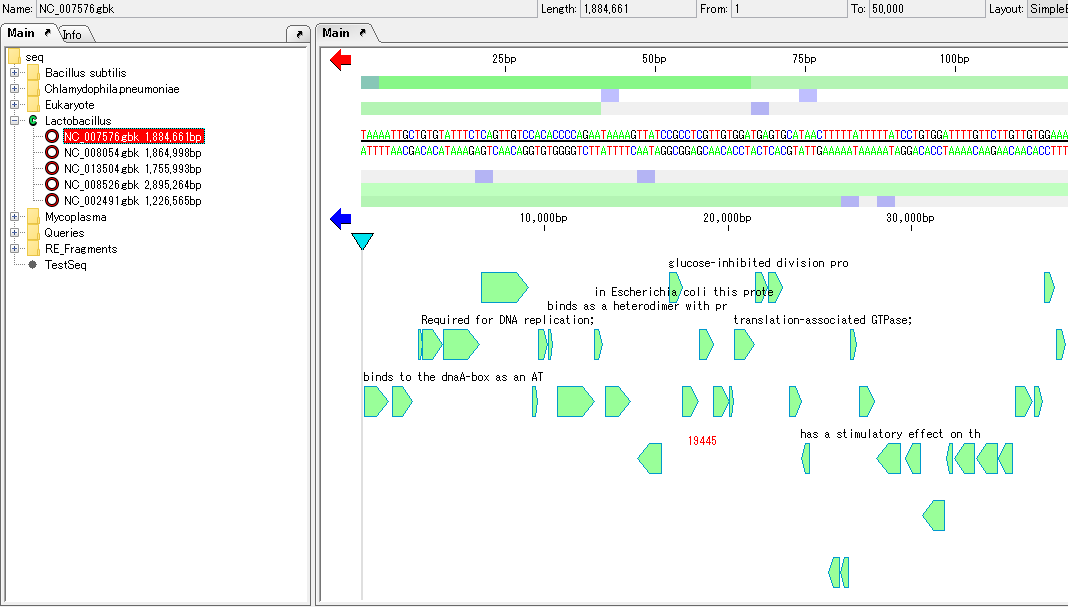
- It works in conjunction with the main feature map, and when you click a feature on the map, it shows where the feature is described.
- Conversely, if you click on any feature description on the GenBank / EMBL viewer, the main feature map will move and display it so that the feature is at the center of the map.
- If you click on an arbitrary sequence row, the feature map will move in the same way and display its position.
Operation
- Load GenBank or EMBL format genome base sequence into the current main directory.
- The feature map for that genome sequence is displayed in the main feature map.
- Click View -> GenBank / EMBL Viewer ... from the menu.
- GenBank / EMBL Viewer will be displayed.
- In the Viewer, the contents of the loaded file are displayed as it is.
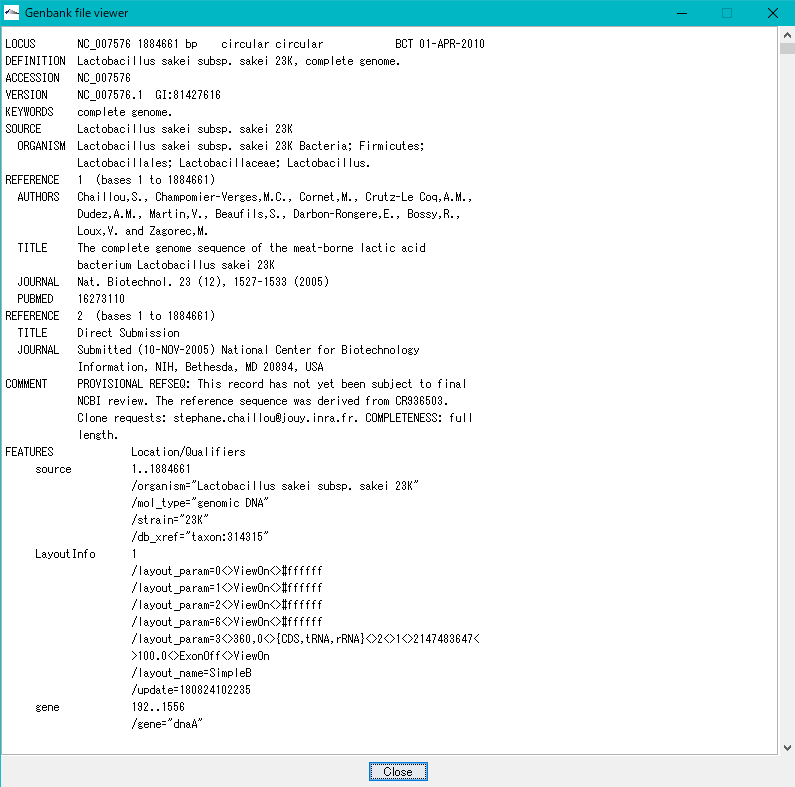
- scroll
- You can scroll vertically by dragging or clicking the scroll bar.
- Move to Feature Position
- Click any feature in GenBank / EMBL Viewer.
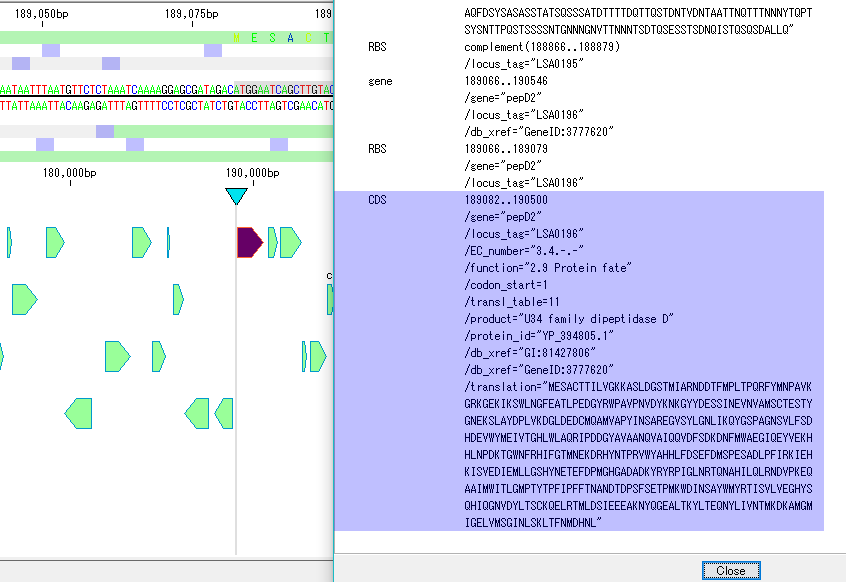
- The description part of that feature is inverted.
- At the same time, the main feature map moves, the feature corresponding to that feature description is displayed centrally and highlighted.
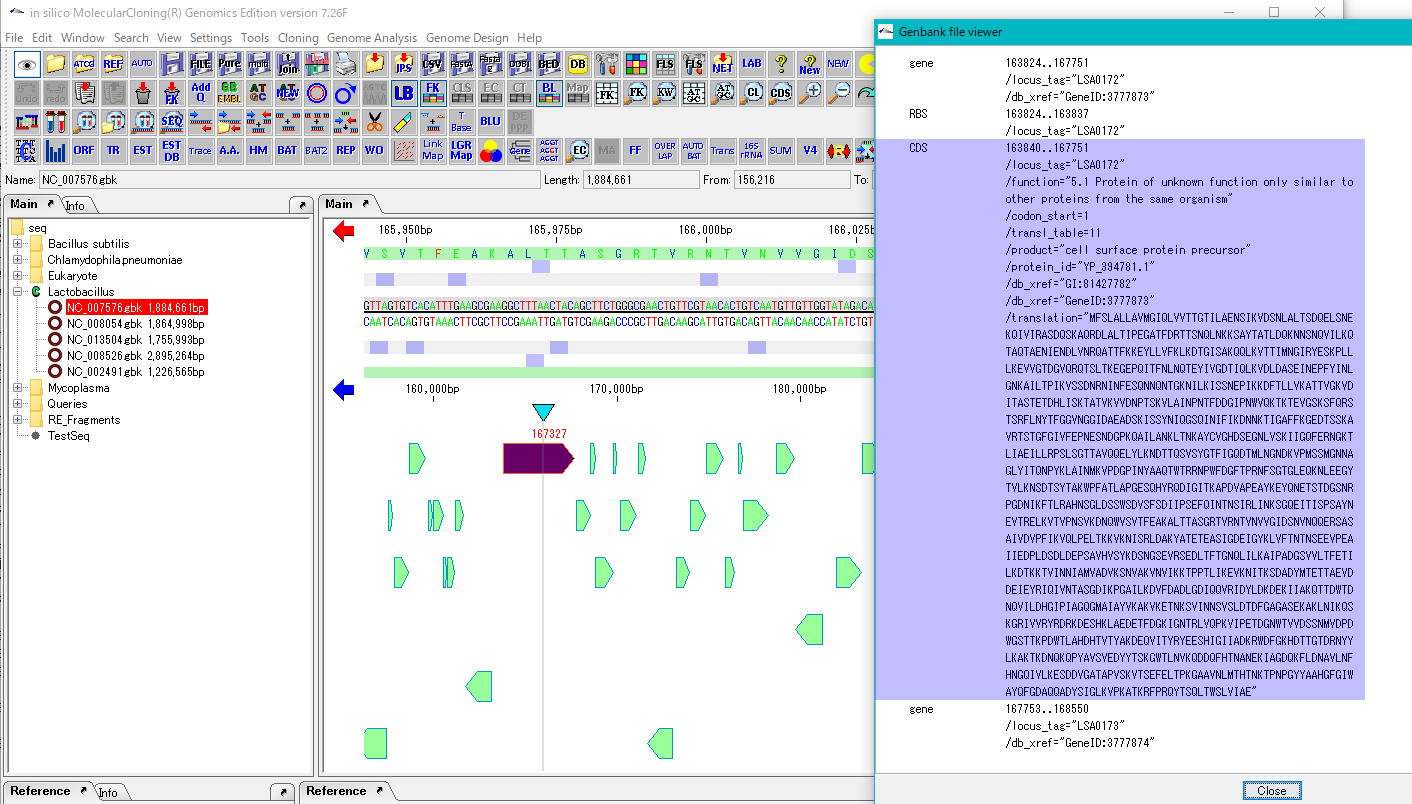
- Conversely, if you click on any feature on the main feature map, GenBank / EMBL Viewer automatically scrolls and its feature description is flipped and displayed.
- Move to sequence position
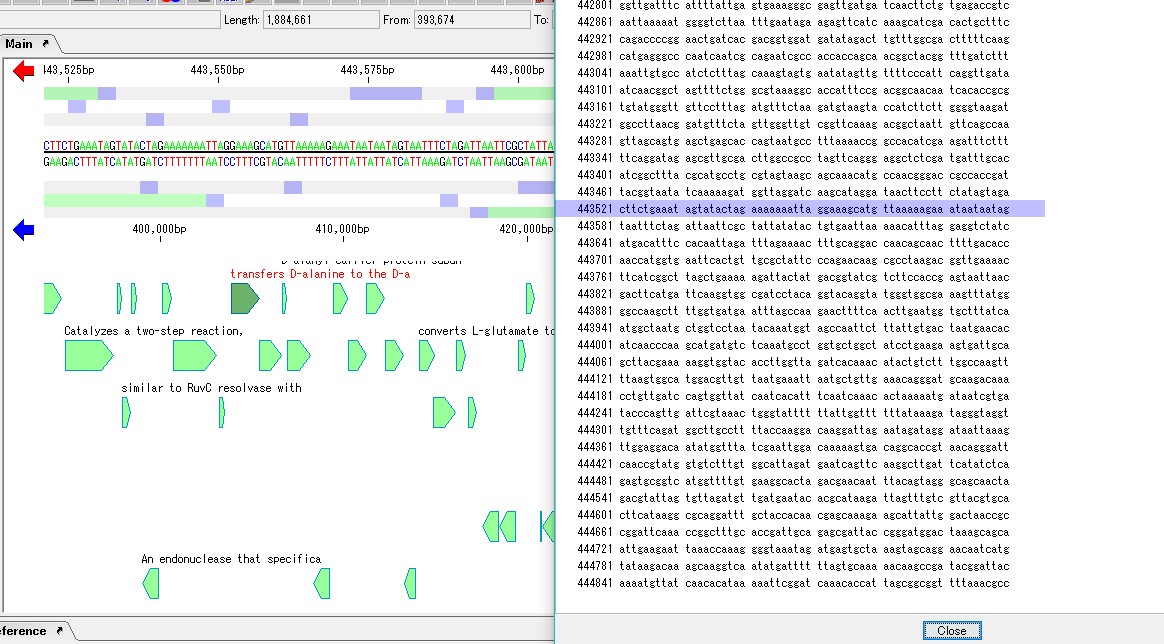
- There is an sequence area in the second half of the GenBank / EMBL format file.
- Clicking on any sequence row will highlight that line and the main feature map will also automatically scroll to show the position of that sequence.
- Clicking any position other than the feature in the main feature map does not move GenBank / EMBL Viewer.
- Closing Viewer
- Click "Close" at the bottom of GenBank / EMBL Viewer to close Viewer.
- You can also close the viewer by changing the current file in the main current directory to another array.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings