IMC W161B Restriction Enzyme Map Lane Setting Dialog Explained
Restriction Enzyme Map Lane Setting Dialog
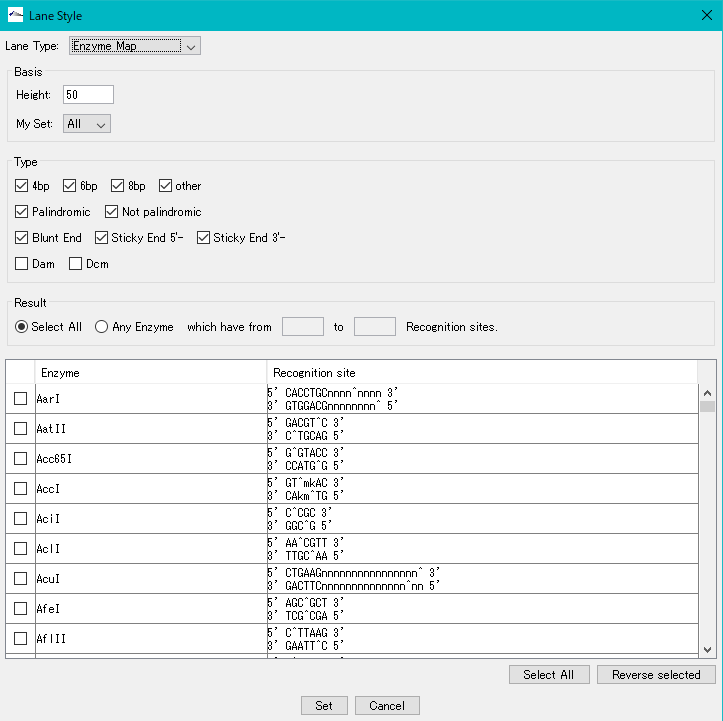
Basis
Height: Set height of restriction enzyme map lane in pixel units
My Set: Select the registered list of restriction enzymes. The default is "All"
Type check box
4 bp check box: check when leaving 4 base recognition restriction enzyme list
6 bp check box: check if leaving 6 base recognition restriction enzyme list
8 bp check box: check when leaving 8 base recognition restriction enzyme on list
Palindromic checkbox: check to leave Palindrome's restriction enzyme list
Not Palindromic check box: check to leave restriction enzymes not on Palindromic list
Blunt End check box: check to keep blunt end restriction enzyme on list
Sticky End 5'-check box: check if the 5 'end leaves a protruding end restriction enzyme list
Sticky End 3'-check box: check if the restriction enzyme whose 3 'end is protruding end is left on the list
DAM check box: check if you want to list the restriction enzymes affected by DAM methylation
DCM checkbox: check if you want to keep the list of restriction enzymes affected by DCM methylation
Result radio button
Select All radio button: Turn on if you do not want to limit by the number of cut parts
Any Enzyme: When restricted to a restriction enzyme having the range of the next cleavage site number, it is on
From: To: Enter the range (positive integer) of the number of recognition sites
Restriction enzyme list
Checkbox: Checked restriction enzyme is targeted
Enzyme: Restriction enzyme name
Recognition Site: recognition site of the restriction enzyme
Select All button: Click to select all restriction enzymes in the restriction enzyme list
Reverse Selected button: Click to reverse the current selection state.
Set button: Click to apply the setting.
Cancel button: Click to cancel the setting.
 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings