- When loading the base sequence file into the current reference directory, the base sequence database for Blast search is automatically generated.
- If the sequence to be loaded is a genomic sequence file CDS is identified and if there is amino acid translation for each CDS, an amino acid sequence search database is generated at the same time.
- In IMCGE, AE, and DS, multiple genome sequences can be loaded simultaneously into the current reference directory, so multiple Blast search databases are generated at the same time.
- The timing for generating the Blast search database is when the new base sequence file is loaded into the current reference directory and when the current reference directory is changed.
- The database of the array which was the current until then is automatically deleted.
- It loads a base sequence file into the current reference directory and automatically generates its base sequence database and amino acid sequence database.
- Make the reference directory that is not the current directory the current reference directory.
- Right click on that directory. The menu will be displayed.
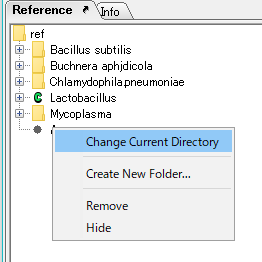
- Select "Change Current Directory" from the menu.
- A confirmation message "Change Current Directory?" Is displayed.
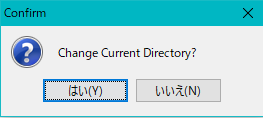
- Click "Yes (Y)". Click "Yes (Y)".
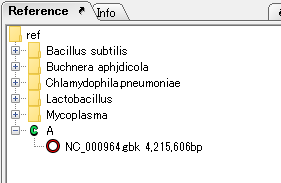
- A directory change will be executed and a green C icon
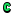 will be displayed in that directory as it completes.
will be displayed in that directory as it completes.
- Load the genomic sequence into the current reference directory.
- From the menu click File -> Load Sequences to MGV.
- The file selection dialog is displayed.
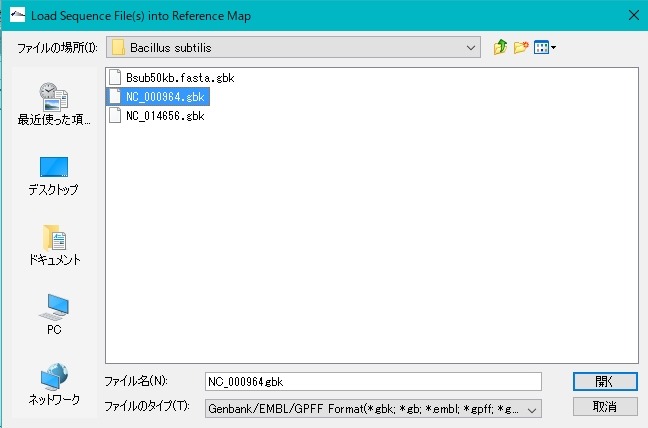
- To use sample array data, select NC_000964.gbk from My Documents> imc_xx> seq> Bacillus subtilis.
- The above genomic base sequence file is loaded into the current reference directory.
- From the menu click File -> Create Blast DB ....
- The "Blast DB List" dialog is displayed.
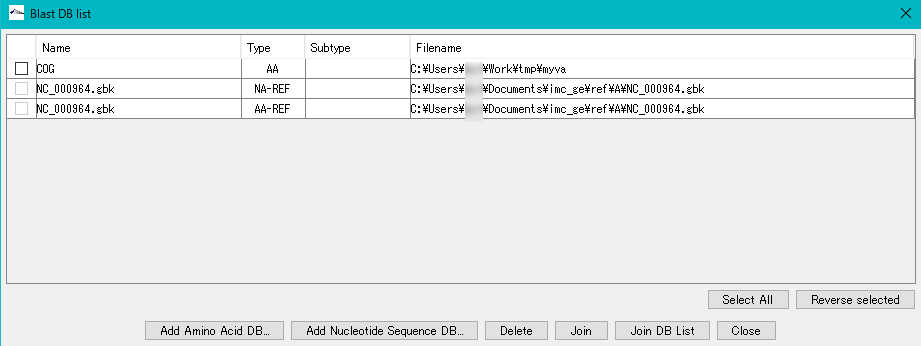
- Two of them are NC - 000964.gbk, and Type is indicated as NA - REF and AA - REF.
- This shows that two types of Blast search databases are generated from the loaded genomic sequence and are automatically registered as a base sequence database and as an amino acid sequence database.


 will be displayed in that directory as it completes.
will be displayed in that directory as it completes.





 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings