- Let's create a network sequence database for executing Blast search on local PC or Mac from amino acid sequence file.
- As an example, we will create a Blast database so that the COG amino acid data file can be searched for in Blast.
- The COG amino acid data file was copied when IMC was installed, and it is saved below.
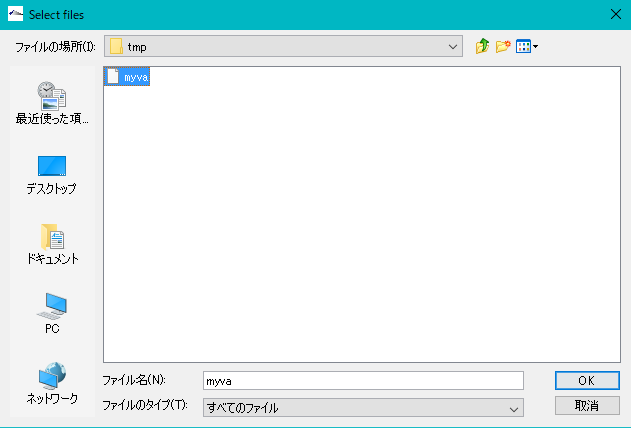
- On Windows, it is stored as myva in c: \ Program Files (x86) \ isb \ imc \ data.
- Copy this file to a temporary storage place in advance. For example, create a temporary directory such as user \ Work and copy it there.
- Operation Start IMC and select File -> Create Blast DB ... from the menu.
-
- The "Blast DB List" dialog is displayed.
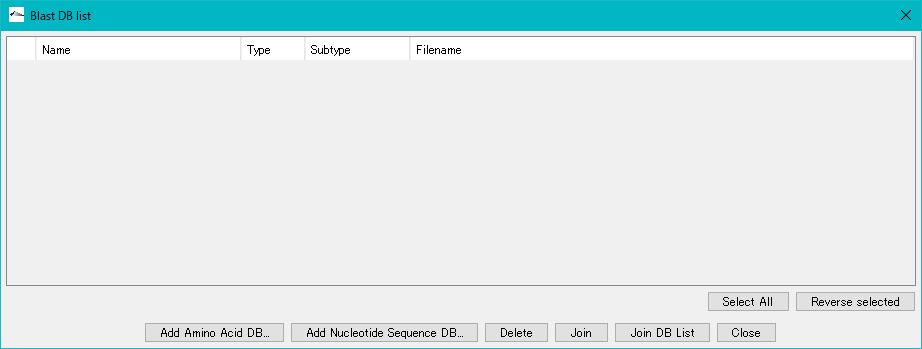
- Click "Add Amino Acid DB ...".
- The "Blast DB Setting" dialog is displayed.
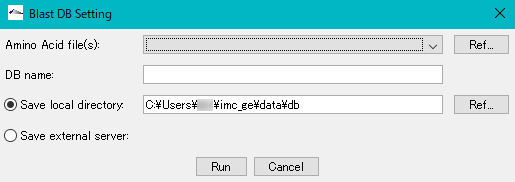
- Amino Acid File (S): Click "Ref ..." on the right side of the column and select the COG file myva that you previously copied.
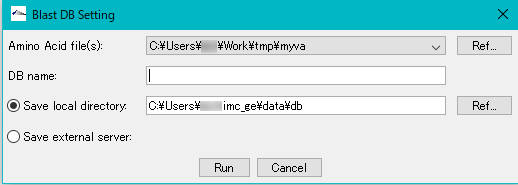
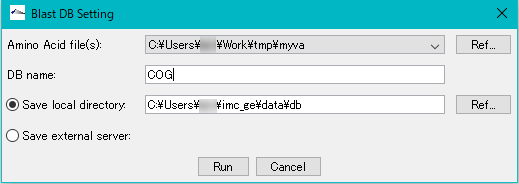
- DB Name: Enter any database name directly in the input field. For example, enter "COG".
- In this time, in order to create a database locally, I have a writable directory of PC or Mac currently in use.
- By default, it is generated in the following directory. In this time, in order to create a database locally, I have a writable directory of PC or Mac currently in use.
- By default, it is generated in the following directory.
- C: \ Users \ user \ imc_xx \ data \ db
- Click "Run".
- A confirmation message "Create Amino Acid DB?" Is displayed.
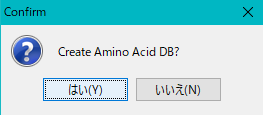
- Click "Yes (Y)". Click "Yes (Y)".
- Execution of the database generation starts, and a progress message is displayed during execution.
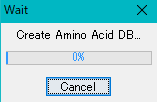
- When generation is completed, a "Completed" message is displayed.
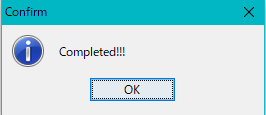
- Clicking "OK" will close the "Completed" message and display the database generated in the Blast DB List.
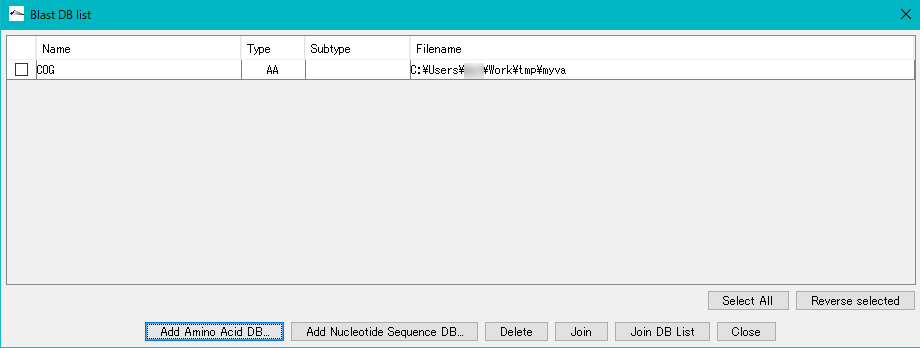
- A database for local blast search has been generated.











 Dongle License (HW Key)
Dongle License (HW Key) Feature Map
Feature Map Management and Operations of Feature Keys
Management and Operations of Feature Keys Sequence and Data Input and Output
Sequence and Data Input and Output GenBank EMBL Viewer
GenBank EMBL Viewer Sequence Viewer
Sequence Viewer Annotation Viewer
Annotation Viewer Circular Genome Viewer-Designer
Circular Genome Viewer-Designer Plasmid Map Viewer-Designer
Plasmid Map Viewer-Designer Trace Viewer - Editor
Trace Viewer - Editor Phylogenetic Tree Viewer
Phylogenetic Tree Viewer Feature Key Search
Feature Key Search Keyword Search
Keyword Search Pattern Search
Pattern Search Priming Site Search
Priming Site Search Batch Homology Search
Batch Homology Search Restriction Enzyme
Restriction Enzyme Primer Design
Primer Design PCR Reaction
PCR Reaction Ligation
Ligation Fragment Modification
Fragment Modification DNA Content Analysis
DNA Content Analysis Codon Analysis
Codon Analysis ORF Analysis
ORF Analysis Database Management
Database Management Multiple Circular Genome Map
Multiple Circular Genome Map Dot Plot Analysis
Dot Plot Analysis Venn Diagram Analysis
Venn Diagram Analysis Reverse Complement
Reverse Complement Settings
Settings